Please be patient as the page loads
|
DBP_HUMAN
|
||||||
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DBP_MOUSE
|
||||||
| θ value | 2.9938e-112 (rank : 1) | NC score | 0.979280 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
DBP_HUMAN
|
||||||
| θ value | 1.3939e-101 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
TEF_MOUSE
|
||||||
| θ value | 4.11246e-53 (rank : 3) | NC score | 0.922514 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
HLF_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 4) | NC score | 0.913407 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
TEF_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 5) | NC score | 0.921942 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
HLF_MOUSE
|
||||||
| θ value | 2.041e-52 (rank : 6) | NC score | 0.912577 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
BATF_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 7) | NC score | 0.416938 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
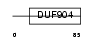
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
BATF_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 8) | NC score | 0.414138 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
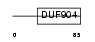
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.170091 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.082037 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.045418 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.316504 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.155112 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
CEBPG_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.343349 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.152790 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.268362 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.075062 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.297293 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPG_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.335230 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |
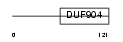
|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.176288 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.062929 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
CEBPD_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.260288 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.164989 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.056144 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.050743 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.102309 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.162338 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.214617 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.035722 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.081573 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.081574 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.077241 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.027856 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
PACN3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.065629 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.031989 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.253295 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.251839 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.038365 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
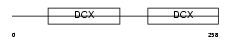
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.026633 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
TMCC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.054034 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80W04, Q6A061 | Gene names | Tmcc2, Kiaa0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2. | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.163805 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.004577 (rank : 155) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.176313 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.171495 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.029549 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.057710 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.049264 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.177941 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.171806 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.029376 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.026948 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.026688 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
DCX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.035281 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
PMYT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.011365 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.156818 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.046744 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.049104 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
GP123_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.035151 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.041716 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TMCC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.049167 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75069, Q6ZN09 | Gene names | TMCC2, KIAA0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2 (Cerebral protein 11). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.142016 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.018505 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.007685 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
NU133_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.039984 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUM0, Q9H9W2, Q9NV71, Q9NVC4 | Gene names | NUP133 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup133 (Nucleoporin Nup133) (133 kDa nucleoporin). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.013814 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.040974 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.030242 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.049635 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.044624 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.044640 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CEBPE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.193121 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.023838 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.007596 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
PACN3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.064714 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99JB8, Q9EQP9 | Gene names | Pacsin3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase II substrate protein 3. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.049722 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.009276 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.030410 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.026382 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.003657 (rank : 156) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.031802 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015488 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.013970 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.021135 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.017092 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MSI2H_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.006076 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.022611 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NKX31_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.004639 (rank : 154) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
SELV_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.060347 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.029071 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.007523 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.007241 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.006896 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.023829 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.020355 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.032666 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
INCE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.014614 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.005891 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.005650 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.024152 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.012494 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.020745 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.020567 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SEP10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.010936 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.005040 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.019142 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.008094 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.014911 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.031551 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.016416 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.014282 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.017842 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GP108_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.011138 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPR9 | Gene names | GPR108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.023924 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.027068 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.002316 (rank : 158) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.035163 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.013697 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.013886 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.032020 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RASF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.015119 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |
|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
ADA33_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.003093 (rank : 157) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.007047 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.039306 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.005851 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.023399 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CU058_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.020913 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.001550 (rank : 159) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.005686 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.052853 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.022693 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.011425 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
STAG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.006258 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.009073 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TMCC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.028923 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.010018 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.005922 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ATF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.136079 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.137146 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.100312 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.110420 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.062552 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.058567 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050843 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CREB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.127676 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.088310 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.092544 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.139373 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.140380 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
JUND_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.069805 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.066553 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054332 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.054457 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.052121 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.069928 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.066035 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.064288 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.078652 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.056181 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.069442 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.3939e-101 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.979280 (rank : 2) | θ value | 2.9938e-112 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.922514 (rank : 3) | θ value | 4.11246e-53 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.921942 (rank : 4) | θ value | 9.16162e-53 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
HLF_HUMAN
|
||||||
| NC score | 0.913407 (rank : 5) | θ value | 9.16162e-53 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| NC score | 0.912577 (rank : 6) | θ value | 2.041e-52 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.416938 (rank : 7) | θ value | 7.1131e-05 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.414138 (rank : 8) | θ value | 0.000158464 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
CEBPG_MOUSE
|
||||||
| NC score | 0.343349 (rank : 9) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CEBPG_HUMAN
|
||||||
| NC score | 0.335230 (rank : 10) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.316504 (rank : 11) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.297293 (rank : 12) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.268362 (rank : 13) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPD_MOUSE
|
||||||
| NC score | 0.260288 (rank : 14) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.253295 (rank : 15) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.251839 (rank : 16) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.214617 (rank : 17) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPE_MOUSE
|
||||||
| NC score | 0.193121 (rank : 18) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.177941 (rank : 19) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.176313 (rank : 20) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.176288 (rank : 21) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.171806 (rank : 22) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.171495 (rank : 23) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.170091 (rank : 24) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.164989 (rank : 25) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.163805 (rank : 26) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.162338 (rank : 27) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.156818 (rank : 28) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.155112 (rank : 29) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.152790 (rank : 30) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.142016 (rank : 31) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.140380 (rank : 32) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.139373 (rank : 33) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.137146 (rank : 34) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.136079 (rank : 35) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.127676 (rank : 36) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.110420 (rank : 37) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.102309 (rank : 38) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.100312 (rank : 39) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.092544 (rank : 40) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.088310 (rank : 41) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.082037 (rank : 42) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.081574 (rank : 43) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.081573 (rank : 44) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.078652 (rank : 45) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.077241 (rank : 46) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.075062 (rank : 47) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.069928 (rank : 48) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.069805 (rank : 49) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.069442 (rank : 50) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.066553 (rank : 51) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.066035 (rank : 52) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PACN3_HUMAN
|
||||||
| NC score | 0.065629 (rank : 53) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
PACN3_MOUSE
|
||||||
| NC score | 0.064714 (rank : 54) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99JB8, Q9EQP9 | Gene names | Pacsin3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase II substrate protein 3. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.064288 (rank : 55) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.062929 (rank : 56) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.062552 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.060347 (rank : 58) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.058567 (rank : 59) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.057710 (rank : 60) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.056181 (rank : 61) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.056144 (rank : 62) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.054457 (rank : 63) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.054332 (rank : 64) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
TMCC2_MOUSE
|
||||||
| NC score | 0.054034 (rank : 65) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80W04, Q6A061 | Gene names | Tmcc2, Kiaa0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.052853 (rank : 66) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.052121 (rank : 67) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.050843 (rank : 68) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.050743 (rank : 69) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.049722 (rank : 70) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.049635 (rank : 71) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.049264 (rank : 72) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TMCC2_HUMAN
|
||||||
| NC score | 0.049167 (rank : 73) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75069, Q6ZN09 | Gene names | TMCC2, KIAA0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2 (Cerebral protein 11). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.049104 (rank : 74) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.046744 (rank : 75) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.045418 (rank : 76) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.044640 (rank : 77) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.044624 (rank : 78) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.041716 (rank : 79) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.040974 (rank : 80) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
NU133_HUMAN
|
||||||
| NC score | 0.039984 (rank : 81) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUM0, Q9H9W2, Q9NV71, Q9NVC4 | Gene names | NUP133 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup133 (Nucleoporin Nup133) (133 kDa nucleoporin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.039306 (rank : 82) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.038365 (rank : 83) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.035722 (rank : 84) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.035281 (rank : 85) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.035163 (rank : 86) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
GP123_HUMAN
|
||||||
| NC score | 0.035151 (rank : 87) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.032666 (rank : 88) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.032020 (rank : 89) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.031989 (rank : 90) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.031802 (rank : 91) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.031551 (rank : 92) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.030410 (rank : 93) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.030242 (rank : 94) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.029549 (rank : 95) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.029376 (rank : 96) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.029071 (rank : 97) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
TMCC1_HUMAN
|
||||||
| NC score | 0.028923 (rank : 98) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.027856 (rank : 99) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.027068 (rank : 100) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.026948 (rank : 101) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.026688 (rank : 102) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.026633 (rank : 103) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.026382 (rank : 104) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.024152 (rank : 105) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.023924 (rank : 106) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.023838 (rank : 107) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.023829 (rank : 108) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.023399 (rank : 109) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.022693 (rank : 110) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.022611 (rank : 111) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.021135 (rank : 112) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CU058_HUMAN
|
||||||
| NC score | 0.020913 (rank : 113) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.020745 (rank : 114) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.020567 (rank : 115) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.020355 (rank : 116) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.019142 (rank : 117) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.018505 (rank : 118) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.017842 (rank : 119) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.017092 (rank : 120) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.016416 (rank : 121) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.015488 (rank : 122) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
RASF8_HUMAN
|
||||||
| NC score | 0.015119 (rank : 123) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.014911 (rank : 124) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.014614 (rank : 125) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.014282 (rank : 126) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.013970 (rank : 127) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.013886 (rank : 128) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.013814 (rank : 129) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.013697 (rank : 130) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.012494 (rank : 131) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.011425 (rank : 132) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PMYT1_MOUSE
|
||||||
| NC score | 0.011365 (rank : 133) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
GP108_HUMAN
|
||||||
| NC score | 0.011138 (rank : 134) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPR9 | Gene names | GPR108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor. | |||||
|
SEP10_HUMAN
|
||||||
| NC score | 0.010936 (rank : 135) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.010018 (rank : 136) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.009276 (rank : 137) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.009073 (rank : 138) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.008094 (rank : 139) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.007685 (rank : 140) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.007596 (rank : 141) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.007523 (rank : 142) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.007241 (rank : 143) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.007047 (rank : 144) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.006896 (rank : 145) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
STAG1_HUMAN
|
||||||
| NC score | 0.006258 (rank : 146) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
MSI2H_HUMAN
|
||||||
| NC score | 0.006076 (rank : 147) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.005922 (rank : 148) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.005891 (rank : 149) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.005851 (rank : 150) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.005686 (rank : 151) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.005650 (rank : 152) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.005040 (rank : 153) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
NKX31_MOUSE
|
||||||
| NC score | 0.004639 (rank : 154) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.004577 (rank : 155) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.003657 (rank : 156) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ADA33_MOUSE
|
||||||
| NC score | 0.003093 (rank : 157) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.002316 (rank : 158) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.001550 (rank : 159) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||