Please be patient as the page loads
|
ATF5_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATF5_HUMAN
|
||||||
| θ value | 7.95186e-81 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | 1.15091e-71 (rank : 2) | NC score | 0.908225 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 3) | NC score | 0.645471 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 4) | NC score | 0.637572 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 5) | NC score | 0.292354 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 6) | NC score | 0.289592 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 7) | NC score | 0.127177 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.081487 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.060665 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.193391 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.195792 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.042989 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.081843 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.177809 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.176548 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.161671 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.178806 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.179071 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.078393 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.071156 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.087810 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.157222 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.075481 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.169666 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.043734 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.065202 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.074857 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HSF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.060529 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
JUND_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.169928 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.045640 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.040013 (rank : 144) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.007436 (rank : 179) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.025371 (rank : 153) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.042823 (rank : 138) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.052168 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.073897 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.073989 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.042097 (rank : 140) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.040205 (rank : 142) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.053786 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
A4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.043694 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.052591 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.051395 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.033975 (rank : 147) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
XKR6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.024429 (rank : 154) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5GH73, Q8TBA0 | Gene names | XKR6, C8orf21, C8orf7, XRG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 6. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.010745 (rank : 174) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.073121 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.074160 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HSF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.047577 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.040179 (rank : 143) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.034260 (rank : 146) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.011751 (rank : 173) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.021434 (rank : 158) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DEDD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.047139 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.006123 (rank : 180) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.005705 (rank : 181) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.040209 (rank : 141) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.024029 (rank : 156) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.064449 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
TPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.032706 (rank : 149) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.019128 (rank : 159) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
ANKZ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.023769 (rank : 157) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.049757 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.045785 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.033802 (rank : 148) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
STRC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.024104 (rank : 155) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
CAR10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.017853 (rank : 163) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.025567 (rank : 152) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.095141 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.035654 (rank : 145) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.031852 (rank : 150) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
FOS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.135207 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.135047 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.070833 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.009068 (rank : 175) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.141737 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.140674 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
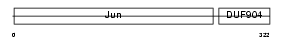
|
|||||
| Description | Transcription factor jun-B. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.096432 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.042594 (rank : 139) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.050049 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.008363 (rank : 178) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
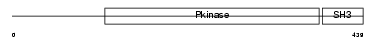
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.014726 (rank : 168) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.026969 (rank : 151) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.049394 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.015242 (rank : 167) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.017863 (rank : 162) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.008995 (rank : 176) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.008621 (rank : 177) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.012627 (rank : 172) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.017756 (rank : 164) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
GAK7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.067622 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
INCE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.013106 (rank : 171) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.014199 (rank : 170) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.015521 (rank : 166) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
IRF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.018123 (rank : 160) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |
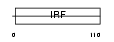
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.005158 (rank : 182) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PYGO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.017910 (rank : 161) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.014652 (rank : 169) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.016259 (rank : 165) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.055060 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.152063 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.151876 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
BATF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.168990 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
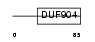
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
BATF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.180653 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054663 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.064521 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055393 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.055955 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051065 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.073630 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |
|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
CEBPG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.073008 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061590 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.063246 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.072188 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.056436 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CO039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056960 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.057503 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.059087 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.064517 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.153440 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.056070 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
DBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.062552 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
DBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.064554 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.058906 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.124372 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.127527 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.068317 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.056132 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058167 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.062616 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.063020 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.066572 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.060362 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.068652 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.067153 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.062950 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054006 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.050379 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.068014 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.059974 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050803 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.060442 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.061159 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.068256 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.067868 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.085674 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058476 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.085704 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.082481 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055422 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054758 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.064398 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.060415 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RU1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.059484 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.058108 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.066303 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.060653 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.057781 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057389 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.056344 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.060256 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.052955 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.054597 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.064508 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.067662 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.066747 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
WASF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.059414 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.057587 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.052992 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.079857 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.083722 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.055980 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.061464 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.054093 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.052375 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.068893 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.065711 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053274 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.062718 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.055112 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.058272 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.065346 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.95186e-81 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.908225 (rank : 2) | θ value | 1.15091e-71 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.645471 (rank : 3) | θ value | 1.47631e-18 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.637572 (rank : 4) | θ value | 3.28887e-18 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.292354 (rank : 5) | θ value | 7.1131e-05 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.289592 (rank : 6) | θ value | 7.1131e-05 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.195792 (rank : 7) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.193391 (rank : 8) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.180653 (rank : 9) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.179071 (rank : 10) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.178806 (rank : 11) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.177809 (rank : 12) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.176548 (rank : 13) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.169928 (rank : 14) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.169666 (rank : 15) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.168990 (rank : 16) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.161671 (rank : 17) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.157222 (rank : 18) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.153440 (rank : 19) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.152063 (rank : 20) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.151876 (rank : 21) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.141737 (rank : 22) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.140674 (rank : 23) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.135207 (rank : 24) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.135047 (rank : 25) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.127527 (rank : 26) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.127177 (rank : 27) | θ value | 0.00175202 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.124372 (rank : 28) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.096432 (rank : 29) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.095141 (rank : 30) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.087810 (rank : 31) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.085704 (rank : 32) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.085674 (rank : 33) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.083722 (rank : 34) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.082481 (rank : 35) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.081843 (rank : 36) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.081487 (rank : 37) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.079857 (rank : 38) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.078393 (rank : 39) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.075481 (rank : 40) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.074857 (rank : 41) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.074160 (rank : 42) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.073989 (rank : 43) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.073897 (rank : 44) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CEBPG_HUMAN
|
||||||
| NC score | 0.073630 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.073121 (rank : 46) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CEBPG_MOUSE
|
||||||
| NC score | 0.073008 (rank : 47) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.072188 (rank : 48) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.071156 (rank : 49) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.070833 (rank : 50) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.068893 (rank : 51) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.068652 (rank : 52) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.068317 (rank : 53) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.068256 (rank : 54) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.068014 (rank : 55) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.067868 (rank : 56) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.067662 (rank : 57) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.067622 (rank : 58) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.067153 (rank : 59) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.066747 (rank : 60) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.066572 (rank : 61) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.066303 (rank : 62) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.065711 (rank : 63) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.065346 (rank : 64) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.065202 (rank : 65) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.064554 (rank : 66) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.064521 (rank : 67) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.064517 (rank : 68) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.064508 (rank : 69) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.064449 (rank : 70) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.064398 (rank : 71) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.063246 (rank : 72) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
GAK17_HUMAN
|
||||||
| NC score | 0.063020 (rank : 73) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK9_HUMAN
|
||||||
| NC score | 0.062950 (rank : 74) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.062718 (rank : 75) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
GAK15_HUMAN
|
||||||
| NC score | 0.062616 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.062552 (rank : 77) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.061590 (rank : 78) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.061464 (rank : 79) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.061159 (rank : 80) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.060665 (rank : 81) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.060653 (rank : 82) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.060529 (rank : 83) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.060442 (rank : 84) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.060415 (rank : 85) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.060362 (rank : 86) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.060256 (rank : 87) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.059974 (rank : 88) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
RU1C_HUMAN
|
||||||
| NC score | 0.059484 (rank : 89) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.059414 (rank : 90) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.059087 (rank : 91) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.058906 (rank : 92) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.058476 (rank : 93) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.058272 (rank : 94) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
GAK14_HUMAN
|
||||||
| NC score | 0.058167 (rank : 95) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
RU1C_MOUSE
|
||||||
| NC score | 0.058108 (rank : 96) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.057781 (rank : 97) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.057587 (rank : 98) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.057503 (rank : 99) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.057389 (rank : 100) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.056960 (rank : 101) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.056436 (rank : 102) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.056344 (rank : 103) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
GAK13_HUMAN
|
||||||
| NC score | 0.056132 (rank : 104) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.056070 (rank : 105) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.055980 (rank : 106) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.055955 (rank : 107) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.055422 (rank : 108) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.055393 (rank : 109) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.055112 (rank : 110) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.055060 (rank : 111) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.054758 (rank : 112) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.054663 (rank : 113) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.054597 (rank : 114) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.054093 (rank : 115) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.054006 (rank : 116) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.053786 (rank : 117) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.053274 (rank : 118) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.052992 (rank : 119) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.052955 (rank : 120) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.052591 (rank : 121) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.052375 (rank : 122) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.052168 (rank : 123) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.051395 (rank : 124) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.051065 (rank : 125) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.050803 (rank : 126) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.050379 (rank : 127) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.050049 (rank : 128) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.049757 (rank : 129) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.049394 (rank : 130) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
HSF4_MOUSE
|
||||||
| NC score | 0.047577 (rank : 131) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
DEDD_MOUSE
|
||||||
| NC score | 0.047139 (rank : 132) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.045785 (rank : 133) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.045640 (rank : 134) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.043734 (rank : 135) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.043694 (rank : 136) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.042989 (rank : 137) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.042823 (rank : 138) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.042594 (rank : 139) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.042097 (rank : 140) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.040209 (rank : 141) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.040205 (rank : 142) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.040179 (rank : 143) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.040013 (rank : 144) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.035654 (rank : 145) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.034260 (rank : 146) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.033975 (rank : 147) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.033802 (rank : 148) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.032706 (rank : 149) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.031852 (rank : 150) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.026969 (rank : 151) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.025567 (rank : 152) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.025371 (rank : 153) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
XKR6_HUMAN
|
||||||
| NC score | 0.024429 (rank : 154) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5GH73, Q8TBA0 | Gene names | XKR6, C8orf21, C8orf7, XRG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 6. | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.024104 (rank : 155) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.024029 (rank : 156) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
ANKZ1_MOUSE
|
||||||
| NC score | 0.023769 (rank : 157) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.021434 (rank : 158) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.019128 (rank : 159) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
IRF2_MOUSE
|
||||||
| NC score | 0.018123 (rank : 160) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
PYGO1_HUMAN
|
||||||
| NC score | 0.017910 (rank : 161) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.017863 (rank : 162) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CAR10_MOUSE
|
||||||
| NC score | 0.017853 (rank : 163) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.017756 (rank : 164) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.016259 (rank : 165) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.015521 (rank : 166) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.015242 (rank : 167) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.014726 (rank : 168) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.014652 (rank : 169) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.014199 (rank : 170) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.013106 (rank : 171) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.012627 (rank : 172) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.011751 (rank : 173) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.010745 (rank : 174) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.009068 (rank : 175) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.008995 (rank : 176) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.008621 (rank : 177) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.008363 (rank : 178) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.007436 (rank : 179) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.006123 (rank : 180) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.005705 (rank : 181) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.005158 (rank : 182) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||