Please be patient as the page loads
|
DEDD_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DEDD_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 2.48205e-183 (rank : 2) | NC score | 0.994928 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
DEDD2_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 3) | NC score | 0.949690 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF8, Q8NBR2, Q8NES1, Q8TAA8, Q96D35 | Gene names | DEDD2, FLAME3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
DEDD2_MOUSE
|
||||||
| θ value | 2.10232e-65 (rank : 4) | NC score | 0.950197 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
ADRB3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.018431 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13945, Q4JFT4 | Gene names | ADRB3, ADRB3R, B3AR | |||
|
Domain Architecture |
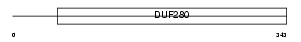
|
|||||
| Description | Beta-3 adrenergic receptor (Beta-3 adrenoceptor) (Beta-3 adrenoreceptor). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.034649 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.056335 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
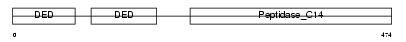
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.031464 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.033323 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
ACHA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.016183 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
B3GA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.032387 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59270 | Gene names | B3gat2, Glcats | |||
|
Domain Architecture |
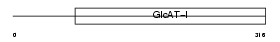
|
|||||
| Description | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 (EC 2.4.1.135) (Beta-1,3-glucuronyltransferase 2) (Glucuronosyltransferase-S) (GlcAT-S) (UDP-glucuronosyltransferase-S) (GlcAT-D). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.025290 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.034207 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.031924 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.049328 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
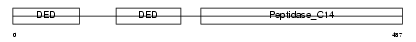
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.033395 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
KLF10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.002945 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89091 | Gene names | Klf10, Gdnfif, Tieg, Tieg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (GDNF-inducible factor) (Transcription factor GIF) (mGIF). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.023636 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.047139 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.012537 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.011826 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.017995 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
TNK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.003430 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.024521 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.060508 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.005232 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.011952 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
NKX28_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.011060 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15522 | Gene names | NKX2-8, NKX-2.8, NKX2G, NKX2H | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.024227 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
DEDD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.994928 (rank : 2) | θ value | 2.48205e-183 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
DEDD2_MOUSE
|
||||||
| NC score | 0.950197 (rank : 3) | θ value | 2.10232e-65 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
DEDD2_HUMAN
|
||||||
| NC score | 0.949690 (rank : 4) | θ value | 1.2325e-65 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXF8, Q8NBR2, Q8NES1, Q8TAA8, Q96D35 | Gene names | DEDD2, FLAME3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.060508 (rank : 5) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.056335 (rank : 6) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.049328 (rank : 7) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.047139 (rank : 8) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.034649 (rank : 9) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.034207 (rank : 10) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.033395 (rank : 11) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.033323 (rank : 12) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
B3GA2_MOUSE
|
||||||
| NC score | 0.032387 (rank : 13) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59270 | Gene names | B3gat2, Glcats | |||
|
Domain Architecture |

|
|||||
| Description | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 (EC 2.4.1.135) (Beta-1,3-glucuronyltransferase 2) (Glucuronosyltransferase-S) (GlcAT-S) (UDP-glucuronosyltransferase-S) (GlcAT-D). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.031924 (rank : 14) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.031464 (rank : 15) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.025290 (rank : 16) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.024521 (rank : 17) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.024227 (rank : 18) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.023636 (rank : 19) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ADRB3_HUMAN
|
||||||
| NC score | 0.018431 (rank : 20) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13945, Q4JFT4 | Gene names | ADRB3, ADRB3R, B3AR | |||
|
Domain Architecture |

|
|||||
| Description | Beta-3 adrenergic receptor (Beta-3 adrenoceptor) (Beta-3 adrenoreceptor). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.017995 (rank : 21) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ACHA4_HUMAN
|
||||||
| NC score | 0.016183 (rank : 22) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.012537 (rank : 23) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.011952 (rank : 24) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.011826 (rank : 25) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
NKX28_HUMAN
|
||||||
| NC score | 0.011060 (rank : 26) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15522 | Gene names | NKX2-8, NKX-2.8, NKX2G, NKX2H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.005232 (rank : 27) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
TNK1_MOUSE
|
||||||
| NC score | 0.003430 (rank : 28) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
KLF10_MOUSE
|
||||||
| NC score | 0.002945 (rank : 29) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89091 | Gene names | Klf10, Gdnfif, Tieg, Tieg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (GDNF-inducible factor) (Transcription factor GIF) (mGIF). | |||||