Please be patient as the page loads
|
CASP8_MOUSE
|
||||||
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
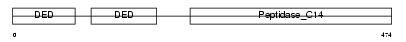
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CASP8_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 1.3161e-184 (rank : 2) | NC score | 0.989068 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
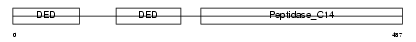
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 5.35859e-61 (rank : 3) | NC score | 0.946179 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
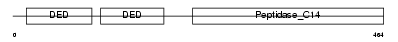
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | 4.55743e-44 (rank : 4) | NC score | 0.913282 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 5) | NC score | 0.909666 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 6) | NC score | 0.905899 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 7) | NC score | 0.906290 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 8) | NC score | 0.888836 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CFLAR_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 9) | NC score | 0.786718 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
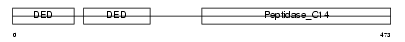
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP9_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 10) | NC score | 0.888690 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 11) | NC score | 0.888813 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 12) | NC score | 0.844745 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 13) | NC score | 0.842098 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
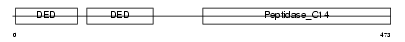
CFLAR_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 14) | NC score | 0.750578 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASPE_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 15) | NC score | 0.810772 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 16) | NC score | 0.809825 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 17) | NC score | 0.647955 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASP1_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 18) | NC score | 0.664192 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
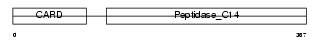
CASP4_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 19) | NC score | 0.654942 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
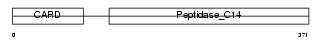
CASP4_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 20) | NC score | 0.620672 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CASP1_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 21) | NC score | 0.648713 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASPC_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 22) | NC score | 0.617864 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
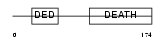
FADD_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 23) | NC score | 0.302427 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 24) | NC score | 0.277227 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
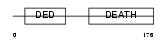
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.187715 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.188735 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
CAR11_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.031902 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.055204 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
DEDD_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.056335 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.022518 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.010993 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.017716 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
VINC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.030183 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |
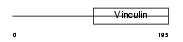
|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
VINC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.028100 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
ARBK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | -0.001317 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25098, Q13837 | Gene names | ADRBK1, BARK, BARK1, GRK2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein coupled receptor kinase 2). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.012755 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.018741 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.011543 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.015149 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.027723 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.009412 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.009948 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.018705 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.010378 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.007940 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.012398 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.013999 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.025364 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.008095 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.008598 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.020383 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
URP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.010763 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006425 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.002358 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.020662 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.021680 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
URP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.010047 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
ICBR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.102279 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |
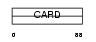
|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.989068 (rank : 2) | θ value | 1.3161e-184 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.946179 (rank : 3) | θ value | 5.35859e-61 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.913282 (rank : 4) | θ value | 4.55743e-44 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.909666 (rank : 5) | θ value | 1.91475e-42 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.906290 (rank : 6) | θ value | 7.52953e-39 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.905899 (rank : 7) | θ value | 8.89434e-40 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.888836 (rank : 8) | θ value | 1.24399e-33 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.888813 (rank : 9) | θ value | 4.00176e-32 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.888690 (rank : 10) | θ value | 1.0531e-32 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.844745 (rank : 11) | θ value | 5.40856e-29 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.842098 (rank : 12) | θ value | 3.50572e-28 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.810772 (rank : 13) | θ value | 2.5182e-18 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.809825 (rank : 14) | θ value | 5.60996e-18 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.786718 (rank : 15) | θ value | 6.17384e-33 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.750578 (rank : 16) | θ value | 2.27234e-27 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.664192 (rank : 17) | θ value | 2.28291e-11 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.654942 (rank : 18) | θ value | 8.67504e-11 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.648713 (rank : 19) | θ value | 2.52405e-10 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.647955 (rank : 20) | θ value | 2.69671e-12 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.620672 (rank : 21) | θ value | 1.133e-10 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.617864 (rank : 22) | θ value | 2.36244e-08 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.302427 (rank : 23) | θ value | 9.92553e-07 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.277227 (rank : 24) | θ value | 4.92598e-06 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.188735 (rank : 25) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.187715 (rank : 26) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
ICBR_HUMAN
|
||||||
| NC score | 0.102279 (rank : 27) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
DEDD_MOUSE
|
||||||
| NC score | 0.056335 (rank : 28) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.055204 (rank : 29) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
CAR11_HUMAN
|
||||||
| NC score | 0.031902 (rank : 30) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
VINC_HUMAN
|
||||||
| NC score | 0.030183 (rank : 31) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
VINC_MOUSE
|
||||||
| NC score | 0.028100 (rank : 32) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.027723 (rank : 33) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.025364 (rank : 34) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.022518 (rank : 35) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.021680 (rank : 36) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.020662 (rank : 37) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.020383 (rank : 38) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.018741 (rank : 39) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.018705 (rank : 40) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.017716 (rank : 41) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.015149 (rank : 42) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.013999 (rank : 43) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.012755 (rank : 44) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.012398 (rank : 45) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.011543 (rank : 46) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.010993 (rank : 47) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
URP2_HUMAN
|
||||||
| NC score | 0.010763 (rank : 48) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.010378 (rank : 49) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
URP2_MOUSE
|
||||||
| NC score | 0.010047 (rank : 50) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.009948 (rank : 51) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.009412 (rank : 52) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.008598 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.008095 (rank : 54) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.007940 (rank : 55) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006425 (rank : 56) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.002358 (rank : 57) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
ARBK1_HUMAN
|
||||||
| NC score | -0.001317 (rank : 58) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25098, Q13837 | Gene names | ADRBK1, BARK, BARK1, GRK2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 1 (EC 2.7.11.15) (Beta-ARK-1) (G- protein coupled receptor kinase 2). | |||||