Please be patient as the page loads
|
CFLAR_HUMAN
|
||||||
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
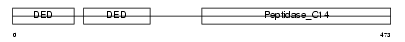
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CFLAR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| θ value | 1.36511e-173 (rank : 2) | NC score | 0.976500 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
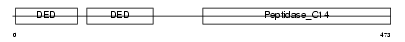
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 3) | NC score | 0.786718 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
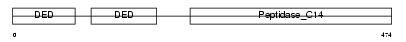
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 4) | NC score | 0.784602 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
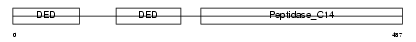
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
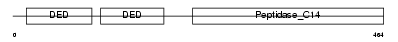
CASPA_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 5) | NC score | 0.752506 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 6) | NC score | 0.658315 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 7) | NC score | 0.655983 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 8) | NC score | 0.625213 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 9) | NC score | 0.649141 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 10) | NC score | 0.622614 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 11) | NC score | 0.651113 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 12) | NC score | 0.640952 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 13) | NC score | 0.651680 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP9_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 14) | NC score | 0.620466 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.441687 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
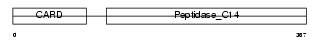
CASP4_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.462670 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 17) | NC score | 0.463917 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASPC_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 18) | NC score | 0.440316 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
CASP1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.464905 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
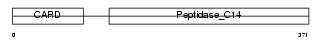
CASP4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 20) | NC score | 0.415509 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.023772 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
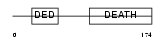
FADD_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.252097 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.010529 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
APOA4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.034210 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06727, Q6Q787 | Gene names | APOA4 | |||
|
Domain Architecture |
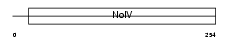
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
MYD88_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.044227 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22366, O35916 | Gene names | Myd88 | |||
|
Domain Architecture |
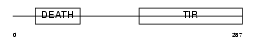
|
|||||
| Description | Myeloid differentiation primary response protein MyD88. | |||||
|
MACOI_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.039916 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MACOI_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.040168 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.010007 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.008810 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.021005 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CASPE_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.485890 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.018462 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
FADD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.196922 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
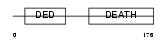
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PP2BC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.015642 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48455 | Gene names | Ppp3cc, Calnc | |||
|
Domain Architecture |
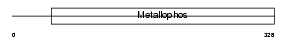
|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.019154 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CASPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.475932 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.013594 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.025509 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
PP2BC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.014286 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48454 | Gene names | PPP3CC, CALNA3, CNA3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
TRI36_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.013705 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.010548 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
LPPRC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.023555 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42704 | Gene names | LRPPRC, LRP130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 130 kDa leucine-rich protein (LRP 130) (GP130) (Leucine-rich PPR motif-containing protein). | |||||
|
BBS7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.015090 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2G4, Q8C7G3, Q8CH00, Q9CXC2 | Gene names | Bbs7, Bbs2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bardet-Biedl syndrome 7 protein homolog (BBS2-like protein 1). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.001351 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CK035_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.015633 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011663 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
TRI36_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.011416 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
ICBR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.067311 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
PEA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.082262 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.083026 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.976500 (rank : 2) | θ value | 1.36511e-173 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.786718 (rank : 3) | θ value | 6.17384e-33 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.784602 (rank : 4) | θ value | 1.0531e-32 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.752506 (rank : 5) | θ value | 1.99067e-23 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.658315 (rank : 6) | θ value | 1.133e-10 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.655983 (rank : 7) | θ value | 1.133e-10 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.651680 (rank : 8) | θ value | 1.38499e-08 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.651113 (rank : 9) | θ value | 2.79066e-09 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.649141 (rank : 10) | θ value | 2.52405e-10 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.640952 (rank : 11) | θ value | 3.64472e-09 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.625213 (rank : 12) | θ value | 1.47974e-10 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.622614 (rank : 13) | θ value | 3.29651e-10 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.620466 (rank : 14) | θ value | 8.40245e-06 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.485890 (rank : 15) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.475932 (rank : 16) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.464905 (rank : 17) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.463917 (rank : 18) | θ value | 0.00035302 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.462670 (rank : 19) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.441687 (rank : 20) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.440316 (rank : 21) | θ value | 0.000786445 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.415509 (rank : 22) | θ value | 0.00869519 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.252097 (rank : 23) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.196922 (rank : 24) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.083026 (rank : 25) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.082262 (rank : 26) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
ICBR_HUMAN
|
||||||
| NC score | 0.067311 (rank : 27) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
MYD88_MOUSE
|
||||||
| NC score | 0.044227 (rank : 28) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22366, O35916 | Gene names | Myd88 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid differentiation primary response protein MyD88. | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.040168 (rank : 29) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.039916 (rank : 30) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
APOA4_HUMAN
|
||||||
| NC score | 0.034210 (rank : 31) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06727, Q6Q787 | Gene names | APOA4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.025509 (rank : 32) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.023772 (rank : 33) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
LPPRC_HUMAN
|
||||||
| NC score | 0.023555 (rank : 34) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42704 | Gene names | LRPPRC, LRP130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 130 kDa leucine-rich protein (LRP 130) (GP130) (Leucine-rich PPR motif-containing protein). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.021005 (rank : 35) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.019154 (rank : 36) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.018462 (rank : 37) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PP2BC_MOUSE
|
||||||
| NC score | 0.015642 (rank : 38) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48455 | Gene names | Ppp3cc, Calnc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
CK035_MOUSE
|
||||||
| NC score | 0.015633 (rank : 39) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
BBS7_MOUSE
|
||||||
| NC score | 0.015090 (rank : 40) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2G4, Q8C7G3, Q8CH00, Q9CXC2 | Gene names | Bbs7, Bbs2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bardet-Biedl syndrome 7 protein homolog (BBS2-like protein 1). | |||||
|
PP2BC_HUMAN
|
||||||
| NC score | 0.014286 (rank : 41) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48454 | Gene names | PPP3CC, CALNA3, CNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
TRI36_HUMAN
|
||||||
| NC score | 0.013705 (rank : 42) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.013594 (rank : 43) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.011663 (rank : 44) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
TRI36_MOUSE
|
||||||
| NC score | 0.011416 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.010548 (rank : 46) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.010529 (rank : 47) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.010007 (rank : 48) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.008810 (rank : 49) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.001351 (rank : 50) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||