Please be patient as the page loads
|
MMP9_HUMAN
|
||||||
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MMP9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990348 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 1.12453e-151 (rank : 3) | NC score | 0.981019 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 1.62381e-150 (rank : 4) | NC score | 0.980949 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 5) | NC score | 0.873476 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | 4.122e-45 (rank : 6) | NC score | 0.873272 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 7) | NC score | 0.875509 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP10_MOUSE
|
||||||
| θ value | 1.56636e-44 (rank : 8) | NC score | 0.873860 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
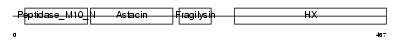
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | 5.95217e-44 (rank : 9) | NC score | 0.874472 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 10) | NC score | 0.872165 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP3_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 11) | NC score | 0.872856 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP8_MOUSE
|
||||||
| θ value | 9.83387e-39 (rank : 12) | NC score | 0.869386 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP12_HUMAN
|
||||||
| θ value | 5.39604e-37 (rank : 13) | NC score | 0.868320 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP1A_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 14) | NC score | 0.868866 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP20_HUMAN
|
||||||
| θ value | 9.20422e-37 (rank : 15) | NC score | 0.868461 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | 1.20211e-36 (rank : 16) | NC score | 0.865862 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP12_MOUSE
|
||||||
| θ value | 3.4976e-36 (rank : 17) | NC score | 0.869912 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 18) | NC score | 0.865027 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP20_MOUSE
|
||||||
| θ value | 6.59618e-35 (rank : 19) | NC score | 0.868196 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 20) | NC score | 0.877205 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
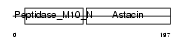
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP8_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 21) | NC score | 0.867041 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP7_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 22) | NC score | 0.877150 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
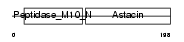
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 23) | NC score | 0.257159 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 24) | NC score | 0.256450 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
MMP1B_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 25) | NC score | 0.865947 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP14_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 26) | NC score | 0.848879 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 27) | NC score | 0.848219 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 28) | NC score | 0.849533 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP24_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 29) | NC score | 0.847625 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP16_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 30) | NC score | 0.845538 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 31) | NC score | 0.845286 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP15_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 32) | NC score | 0.843718 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 33) | NC score | 0.850066 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 34) | NC score | 0.839129 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 35) | NC score | 0.850148 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP26_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 36) | NC score | 0.867968 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
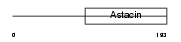
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
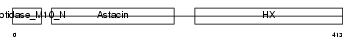
MMP19_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 37) | NC score | 0.843510 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 38) | NC score | 0.850169 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP28_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 39) | NC score | 0.840574 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
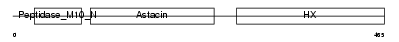
MMP19_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 40) | NC score | 0.838335 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 41) | NC score | 0.270799 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 42) | NC score | 0.276342 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 43) | NC score | 0.395309 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 44) | NC score | 0.390595 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 45) | NC score | 0.261650 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 46) | NC score | 0.262005 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MMP21_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 47) | NC score | 0.794929 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
LY75_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 48) | NC score | 0.250148 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 49) | NC score | 0.290011 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 50) | NC score | 0.093730 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
MPRI_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 51) | NC score | 0.284082 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
FA12_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 52) | NC score | 0.095959 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
MMP21_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 53) | NC score | 0.788658 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 54) | NC score | 0.246667 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
HGFA_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 55) | NC score | 0.092777 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 56) | NC score | 0.613358 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
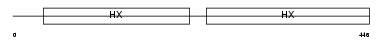
HEMO_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 57) | NC score | 0.536379 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |
|
|||||
| Description | Hemopexin precursor. | |||||
|
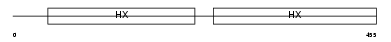
HEMO_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 58) | NC score | 0.579795 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |
|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 59) | NC score | 0.584703 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 60) | NC score | 0.029773 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 61) | NC score | 0.323533 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 62) | NC score | 0.033448 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 63) | NC score | 0.034044 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 64) | NC score | 0.024030 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CP003_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 65) | NC score | 0.092605 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 66) | NC score | 0.022417 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
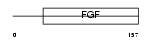
FGF23_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 67) | NC score | 0.024494 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPC2 | Gene names | Fgf23 | |||
|
Domain Architecture |
|
|||||
| Description | Fibroblast growth factor 23 precursor (FGF-23). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 0.125558 (rank : 68) | NC score | 0.016141 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 69) | NC score | 0.025937 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 70) | NC score | 0.034400 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TSNA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 71) | NC score | 0.034967 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 0.279714 (rank : 72) | NC score | 0.049036 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 0.365318 (rank : 73) | NC score | 0.019945 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.019986 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.038278 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
3BHS6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.017034 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.058913 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.009585 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.000013 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.011076 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.019454 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.023117 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.014005 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.016548 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.020753 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.029424 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.012226 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.022354 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.010292 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.005176 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.013306 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024825 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.033237 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.009486 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.006495 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.010886 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.013723 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.004806 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.000873 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.008905 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
RT35_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.010157 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJZ4, Q8VCG7 | Gene names | Mrps35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S35, mitochondrial precursor (S35mt) (MRP-S35). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | -0.003580 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.018537 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.018279 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.016159 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.013990 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
DEDD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.011952 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | -0.006086 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
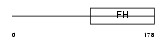
FOXO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.007870 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.013783 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
NKX31_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | -0.002051 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
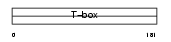
TBX18_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.001847 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.004118 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ADA15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.007352 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.013023 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.000740 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
ESAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.001586 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.010141 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.007324 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.008885 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.041738 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006495 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.003993 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.016960 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.013557 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CD302_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.076323 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.075396 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
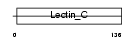
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.060266 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060181 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
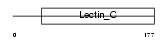
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
EMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.053566 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
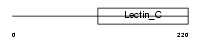
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.054916 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
PRG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052523 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.264892 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.990348 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.981019 (rank : 3) | θ value | 1.12453e-151 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.980949 (rank : 4) | θ value | 1.62381e-150 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.877205 (rank : 5) | θ value | 8.61488e-35 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.877150 (rank : 6) | θ value | 1.6247e-33 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.875509 (rank : 7) | θ value | 9.18288e-45 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.874472 (rank : 8) | θ value | 5.95217e-44 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.873860 (rank : 9) | θ value | 1.56636e-44 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.873476 (rank : 10) | θ value | 2.85459e-46 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.873272 (rank : 11) | θ value | 4.122e-45 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.872856 (rank : 12) | θ value | 6.58091e-43 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.872165 (rank : 13) | θ value | 2.95404e-43 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP12_MOUSE
|
||||||
| NC score | 0.869912 (rank : 14) | θ value | 3.4976e-36 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP8_MOUSE
|
||||||
| NC score | 0.869386 (rank : 15) | θ value | 9.83387e-39 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP1A_MOUSE
|
||||||
| NC score | 0.868866 (rank : 16) | θ value | 7.04741e-37 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP20_HUMAN
|
||||||
| NC score | 0.868461 (rank : 17) | θ value | 9.20422e-37 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP12_HUMAN
|
||||||
| NC score | 0.868320 (rank : 18) | θ value | 5.39604e-37 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP20_MOUSE
|
||||||
| NC score | 0.868196 (rank : 19) | θ value | 6.59618e-35 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.867968 (rank : 20) | θ value | 1.80048e-24 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP8_HUMAN
|
||||||
| NC score | 0.867041 (rank : 21) | θ value | 9.52487e-34 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP1B_MOUSE
|
||||||
| NC score | 0.865947 (rank : 22) | θ value | 1.37539e-32 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.865862 (rank : 23) | θ value | 1.20211e-36 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.865027 (rank : 24) | θ value | 1.32909e-35 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.850169 (rank : 25) | θ value | 1.09232e-21 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.850148 (rank : 26) | θ value | 5.59698e-26 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.850066 (rank : 27) | θ value | 2.96777e-27 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| NC score | 0.849533 (rank : 28) | θ value | 3.74554e-30 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP14_HUMAN
|
||||||
| NC score | 0.848879 (rank : 29) | θ value | 1.52067e-31 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| NC score | 0.848219 (rank : 30) | θ value | 2.59387e-31 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP24_HUMAN
|
||||||
| NC score | 0.847625 (rank : 31) | θ value | 8.3442e-30 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP16_HUMAN
|
||||||
| NC score | 0.845538 (rank : 32) | θ value | 1.08979e-29 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.845286 (rank : 33) | θ value | 2.42779e-29 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP15_HUMAN
|
||||||
| NC score | 0.843718 (rank : 34) | θ value | 9.2256e-29 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP19_MOUSE
|
||||||
| NC score | 0.843510 (rank : 35) | θ value | 4.43474e-23 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP28_HUMAN
|
||||||
| NC score | 0.840574 (rank : 36) | θ value | 4.15078e-21 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.839129 (rank : 37) | θ value | 3.87602e-27 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP19_HUMAN
|
||||||
| NC score | 0.838335 (rank : 38) | θ value | 7.32683e-18 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP21_HUMAN
|
||||||
| NC score | 0.794929 (rank : 39) | θ value | 6.00763e-12 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP21_MOUSE
|
||||||
| NC score | 0.788658 (rank : 40) | θ value | 3.64472e-09 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.613358 (rank : 41) | θ value | 1.29631e-06 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.584703 (rank : 42) | θ value | 0.000602161 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
HEMO_HUMAN
|
||||||
| NC score | 0.579795 (rank : 43) | θ value | 5.44631e-05 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
HEMO_MOUSE
|
||||||
| NC score | 0.536379 (rank : 44) | θ value | 3.19293e-05 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor. | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.395309 (rank : 45) | θ value | 5.8054e-15 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.390595 (rank : 46) | θ value | 5.8054e-15 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.323533 (rank : 47) | θ value | 0.000786445 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.290011 (rank : 48) | θ value | 4.30538e-10 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MPRI_HUMAN
|
||||||
| NC score | 0.284082 (rank : 49) | θ value | 2.13673e-09 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.276342 (rank : 50) | θ value | 1.52774e-15 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.270799 (rank : 51) | θ value | 6.85773e-16 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.264892 (rank : 52) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.262005 (rank : 53) | θ value | 1.09485e-13 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.261650 (rank : 54) | θ value | 3.76295e-14 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.257159 (rank : 55) | θ value | 2.12192e-33 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.256450 (rank : 56) | θ value | 6.17384e-33 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.250148 (rank : 57) | θ value | 2.52405e-10 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.246667 (rank : 58) | θ value | 6.21693e-09 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.095959 (rank : 59) | θ value | 3.64472e-09 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.093730 (rank : 60) | θ value | 9.59137e-10 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.092777 (rank : 61) | θ value | 1.38499e-08 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
CP003_HUMAN
|
||||||
| NC score | 0.092605 (rank : 62) | θ value | 0.0252991 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.076323 (rank : 63) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.075396 (rank : 64) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.060266 (rank : 65) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.060181 (rank : 66) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.058913 (rank : 67) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.054916 (rank : 68) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.053566 (rank : 69) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.052523 (rank : 70) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.049036 (rank : 71) | θ value | 0.279714 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.041738 (rank : 72) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.038278 (rank : 73) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TSNA1_HUMAN
|
||||||
| NC score | 0.034967 (rank : 74) | θ value | 0.21417 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.034400 (rank : 75) | θ value | 0.163984 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.034044 (rank : 76) | θ value | 0.0148317 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.033448 (rank : 77) | θ value | 0.00665767 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.033237 (rank : 78) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.029773 (rank : 79) | θ value | 0.000786445 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.029424 (rank : 80) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.025937 (rank : 81) | θ value | 0.163984 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.024825 (rank : 82) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
FGF23_MOUSE
|
||||||
| NC score | 0.024494 (rank : 83) | θ value | 0.0563607 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPC2 | Gene names | Fgf23 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor 23 precursor (FGF-23). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.024030 (rank : 84) | θ value | 0.0148317 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.023117 (rank : 85) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.022417 (rank : 86) | θ value | 0.0563607 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.022354 (rank : 87) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.020753 (rank : 88) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.019986 (rank : 89) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.019945 (rank : 90) | θ value | 0.365318 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.019454 (rank : 91) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.018537 (rank : 92) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS2_MOUSE
|
||||||
| NC score | 0.018279 (rank : 93) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
3BHS6_MOUSE
|
||||||
| NC score | 0.017034 (rank : 94) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.016960 (rank : 95) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.016548 (rank : 96) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
ATS3_HUMAN
|
||||||
| NC score | 0.016159 (rank : 97) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.016141 (rank : 98) | θ value | 0.125558 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.014005 (rank : 99) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.013990 (rank : 100) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.013783 (rank : 101) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.013723 (rank : 102) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.013557 (rank : 103) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.013306 (rank : 104) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.013023 (rank : 105) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.012226 (rank : 106) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DEDD_MOUSE
|
||||||
| NC score | 0.011952 (rank : 107) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.011076 (rank : 108) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.010886 (rank : 109) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.010292 (rank : 110) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
RT35_MOUSE
|
||||||
| NC score | 0.010157 (rank : 111) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJZ4, Q8VCG7 | Gene names | Mrps35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S35, mitochondrial precursor (S35mt) (MRP-S35). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.010141 (rank : 112) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.009585 (rank : 113) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.009486 (rank : 114) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.008905 (rank : 115) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.008885 (rank : 116) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.007870 (rank : 117) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
ADA15_MOUSE
|
||||||
| NC score | 0.007352 (rank : 118) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.007324 (rank : 119) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.006495 (rank : 120) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.006495 (rank : 121) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.005176 (rank : 122) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.004806 (rank : 123) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.004118 (rank : 124) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.003993 (rank : 125) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.001847 (rank : 126) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
ESAM_HUMAN
|
||||||
| NC score | 0.001586 (rank : 127) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.000873 (rank : 128) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.000740 (rank : 129) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.000013 (rank : 130) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
NKX31_MOUSE
|
||||||
| NC score | -0.002051 (rank : 131) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
AAK1_MOUSE
|
||||||
| NC score | -0.003580 (rank : 132) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
EGR2_HUMAN
|
||||||
| NC score | -0.006086 (rank : 133) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||