Please be patient as the page loads
|
PRG3_MOUSE
|
||||||
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRG3_MOUSE
|
||||||
| θ value | 2.51683e-135 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | 4.66183e-81 (rank : 2) | NC score | 0.960865 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
EMBP_MOUSE
|
||||||
| θ value | 1.07972e-61 (rank : 3) | NC score | 0.918159 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
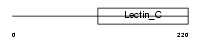
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
EMBP_HUMAN
|
||||||
| θ value | 4.11246e-53 (rank : 4) | NC score | 0.918150 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13727, Q14227 | Gene names | PRG2, MBP | |||
|
Domain Architecture |
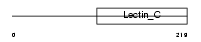
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (EMBP) (Pregnancy-associated major basic protein) (Proteoglycan 2, bone marrow) (BMPG). | |||||
|
MRC1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.489405 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.481569 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
REG1B_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 7) | NC score | 0.512165 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |
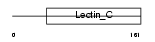
|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 8) | NC score | 0.469878 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
FCER2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 9) | NC score | 0.396678 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 10) | NC score | 0.301170 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 11) | NC score | 0.465657 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 12) | NC score | 0.301563 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
LIT2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 13) | NC score | 0.506115 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |
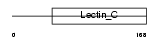
|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
LIT1_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 14) | NC score | 0.506771 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |
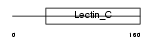
|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 15) | NC score | 0.452104 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
REG1A_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.475384 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |
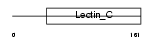
|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG3A_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 17) | NC score | 0.468637 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |
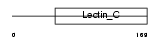
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
REG3G_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 18) | NC score | 0.500865 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |
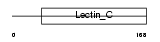
|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
SFTPD_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 19) | NC score | 0.256465 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P50404 | Gene names | Sftpd, Sftp4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
REG3A_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 20) | NC score | 0.485866 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |
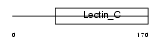
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
LY75_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 21) | NC score | 0.453129 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 22) | NC score | 0.332284 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |
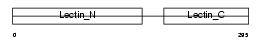
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
REG3G_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 23) | NC score | 0.450250 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 24) | NC score | 0.311271 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.316028 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 26) | NC score | 0.300777 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 27) | NC score | 0.346277 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SFTPA_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 28) | NC score | 0.288557 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35242 | Gene names | Sftpa1, Sftp-1, Sftp1, Sftpa | |||
|
Domain Architecture |
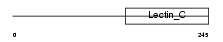
|
|||||
| Description | Pulmonary surfactant-associated protein A precursor (SP-A) (PSP-A) (PSAP). | |||||
|
FCER2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 29) | NC score | 0.412137 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
C209E_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 30) | NC score | 0.312877 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |
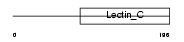
|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.332914 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
SFTPD_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 32) | NC score | 0.248405 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35247, Q86YK9, Q9UCJ2, Q9UCJ3 | Gene names | SFTPD, PSPD, SFTP4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
FREM1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.245646 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
ASGR1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.326733 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
ASGR1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.314460 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 36) | NC score | 0.327470 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
REG3B_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.449951 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
SFTA1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.290034 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
SFTA2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.286887 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
CLC4K_MOUSE
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.305535 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.315606 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.053635 (rank : 138) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.318348 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
FREM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.222317 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.057546 (rank : 135) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.030071 (rank : 153) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.032509 (rank : 150) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ZN646_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.004734 (rank : 184) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
CLC4G_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.357959 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
MBL2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.250540 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |
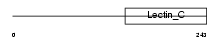
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.038646 (rank : 145) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CLC4K_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.313280 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.051228 (rank : 142) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.031422 (rank : 151) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TETN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.364578 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |
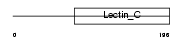
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
REN3B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.023422 (rank : 156) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
TETN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.364094 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |
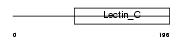
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.026013 (rank : 155) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
DPOE3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.034542 (rank : 148) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |
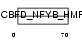
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.022436 (rank : 159) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.042115 (rank : 144) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.005037 (rank : 183) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.034570 (rank : 147) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.006466 (rank : 179) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.014540 (rank : 168) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
NFL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.007914 (rank : 175) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.015930 (rank : 166) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.032902 (rank : 149) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.022682 (rank : 158) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.021613 (rank : 160) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.049240 (rank : 143) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
D2HDH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.022868 (rank : 157) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIM3, Q3TDF5, Q8BU06 | Gene names | D2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
LYAM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.133713 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.006953 (rank : 177) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.016959 (rank : 165) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.013110 (rank : 169) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CCD27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.009774 (rank : 173) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
AN13A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.007816 (rank : 176) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UP5, Q2VPQ7, Q6P7F2 | Gene names | Ankrd13a, Ankrd13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13A. | |||||
|
CLC4G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.326004 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.008012 (rank : 174) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.006309 (rank : 180) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009999 (rank : 172) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | -0.002417 (rank : 188) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.031151 (rank : 152) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
FHL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.001907 (rank : 186) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.019292 (rank : 162) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.053099 (rank : 140) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.006679 (rank : 178) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.035858 (rank : 146) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.018060 (rank : 163) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.005162 (rank : 182) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.015416 (rank : 167) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.017363 (rank : 164) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | -0.002037 (rank : 187) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.010280 (rank : 170) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | -0.002633 (rank : 189) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD302_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.233523 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.335925 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
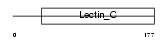
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.010164 (rank : 171) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.004215 (rank : 185) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.026154 (rank : 154) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.019443 (rank : 161) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
THYG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.005259 (rank : 181) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
ASGR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.299783 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |
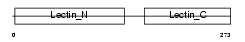
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054599 (rank : 137) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
C209A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.277211 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |
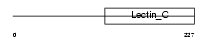
|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
C209B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.264721 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
C209C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.286192 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |
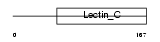
|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
C209D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.277086 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |
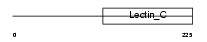
|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
CD209_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.262607 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
CD248_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058726 (rank : 133) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD248_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061564 (rank : 131) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.232387 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CD69_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.206595 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |
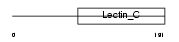
|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CD69_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.194171 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
CD72_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.146816 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P21854 | Gene names | CD72 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2). | |||||
|
CD72_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.064319 (rank : 128) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21855, Q64225 | Gene names | Cd72, Ly-32, Lyb-2 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2) (Antigen Ly-32). | |||||
|
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.336495 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
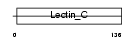
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CLC11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.187062 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |
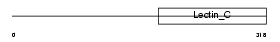
|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
CLC11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.201447 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88200, Q8C946, Q9CTF0 | Gene names | Clec11a, Scgf | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin). | |||||
|
CLC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.150772 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NC01, Q8IUW7, Q9NZH3 | Gene names | CLEC1A, CLEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A (C-type lectin-like receptor 1) (CLEC-1). | |||||
|
CLC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.183553 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BWY2, Q7TPX7, Q8BMH5 | Gene names | Clec1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A. | |||||
|
CLC2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.165254 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92478, Q8IZE9, Q9BS74, Q9UQB4 | Gene names | CLEC2B, AICL, CLECSF2, IFNRG1 | |||
|
Domain Architecture |
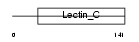
|
|||||
| Description | C-type lectin domain family 2 member B (C-type lectin superfamily member 2) (Activation-induced C-type lectin) (IFN-alpha2b-inducing- related protein 1) (C-type lectin domain family 2 member B). | |||||
|
CLC3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.296957 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |
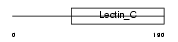
|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.297648 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.255375 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
CLC4A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.241167 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
CLC4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.230811 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
CLC4D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.277989 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
CLC4D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.268223 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
CLC4E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.250208 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.260093 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.136713 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N1N0 | Gene names | CLEC4F, CLECSF13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.214939 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
CLC4M_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.256284 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
CLC5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.143684 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NY25, Q9UKQ0 | Gene names | CLEC5A, CLECSF5, MDL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 5 member A (C-type lectin superfamily member 5) (Myeloid DAP12-associating lectin) (MDL-1). | |||||
|
CLC5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.160860 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R007, Q3UV97, Q8BL24 | Gene names | Clec5a, Clecsf5, Mdl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 5 member A (C-type lectin superfamily member 5) (Myeloid DAP12-associating lectin protein) (MDL-1). | |||||
|
CLC6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.251697 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.242136 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC9A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.170126 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UXN8 | Gene names | CLEC9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
CLC9A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.175436 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRU4 | Gene names | Clec9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
HPLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.131014 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |
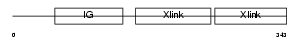
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.130419 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |
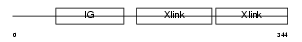
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.129711 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |
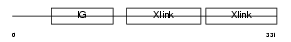
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.130094 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |
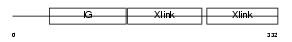
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.132361 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |
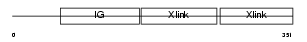
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
HPLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.133782 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |
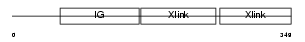
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
HPLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.128852 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |
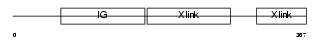
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.127395 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
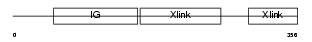
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
IDD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.073765 (rank : 127) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
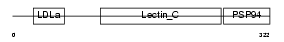
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
IDD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.078156 (rank : 126) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
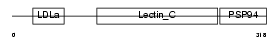
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
KLRA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.116057 (rank : 115) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64329, Q61154, Q64257 | Gene names | Klra3, Ly-49c, Ly49C | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor 3 (T-cell surface glycoprotein Ly- 49C) (Ly49-C antigen) (Lymphocyte antigen 49C) (5E6). | |||||
|
KLRA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.108446 (rank : 118) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60651, O78026, Q9EPA5 | Gene names | Klra4, Ly-49d, Ly49-d, Ly49d | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor 4 (T-cell surface glycoprotein Ly- 49D) (Ly49-D antigen) (Lymphocyte antigen 49D). | |||||
|
KLRA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.111209 (rank : 117) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60652 | Gene names | Klra5, Ly-49e, Ly49-e, Ly49e | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor 5 (T-cell surface glycoprotein Ly- 49E) (Ly49-E antigen) (Lymphocyte antigen 49E). | |||||
|
KLRA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.101191 (rank : 119) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60653 | Gene names | Klra6, Ly-49f, Ly49-f, Ly49F | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor 6 (T-cell surface glycoprotein Ly- 49F) (Ly49-F antigen) (Lymphocyte antigen 49F). | |||||
|
KLRA7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.119393 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60654, Q60655, Q60656, Q60683 | Gene names | Klra7, Ly-49g, Ly49-g, Ly49g, Ly49g4 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 7 (T-cell surface glycoprotein Ly- 49G) (Ly49-G antigen) (Lymphocyte antigen 49G). | |||||
|
KLRA8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.097529 (rank : 122) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60682, O78027 | Gene names | Klra8, Ly-49h, Ly49-h, Ly49H | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor 8 (T-cell surface glycoprotein Ly- 49H) (Ly49-H antigen) (Lymphocyte antigen 49H). | |||||
|
KLRBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.160404 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27811 | Gene names | Klrb1a, Ly55, Ly55a | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-2 (NKR-P1 2) (NKR-P1.7). | |||||
|
KLRBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.181150 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27812 | Gene names | Klrb1b, Ly55-b, Ly55b | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-34 (NKR-P1 34). | |||||
|
KLRBC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.156957 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27814 | Gene names | Klrb1c, Ly55-c, Ly55c | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-40 (NKR-P1 40) (NKR-P1.9). | |||||
|
KLRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.213243 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
KLRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.241561 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
LY49A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.113730 (rank : 116) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20937 | Gene names | Klra1, Ly-49, Ly-49a, Ly49, Ly49A | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein YE1/48 (T lymphocyte antigen A1) (Ly49-A antigen) (Lymphocyte antigen 49A). | |||||
|
LYAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.147901 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.152091 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |
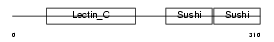
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.118790 (rank : 114) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
LYAM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.100436 (rank : 121) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
LYAM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.094623 (rank : 123) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
MBL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.263767 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |
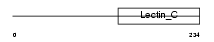
|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
MBL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.255859 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |
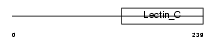
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.052523 (rank : 141) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
NKG2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.168341 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P26715 | Gene names | KLRC1, NKG2A | |||
|
Domain Architecture |
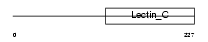
|
|||||
| Description | NKG2-A/NKG2-B type II integral membrane protein (NKG2-A/B-activating NK receptor) (NK cell receptor A) (CD159a antigen). | |||||
|
NKG2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.169864 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26717, O43802, Q9NR42 | Gene names | KLRC2, NKG2C | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-C type II integral membrane protein (NKG2-C-activating NK receptor) (NK cell receptor C) (CD159c antigen). | |||||
|
NKG2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.230101 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
NKG2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.234381 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
NKG2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.173896 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07444, Q96RL0, Q9UP04 | Gene names | KLRC3, NKG2E | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-E type II integral membrane protein (NKG2-E-activating NK receptor) (NK cell receptor E). | |||||
|
OLR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.148821 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
OLR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.126361 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.053619 (rank : 139) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.100748 (rank : 120) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
REG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.389220 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
REG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.367796 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.086348 (rank : 125) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.087439 (rank : 124) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
TRBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.055911 (rank : 136) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.058248 (rank : 134) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.058895 (rank : 132) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
XLKD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.062544 (rank : 130) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.064185 (rank : 129) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.51683e-135 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.960865 (rank : 2) | θ value | 4.66183e-81 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.918159 (rank : 3) | θ value | 1.07972e-61 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
EMBP_HUMAN
|
||||||
| NC score | 0.918150 (rank : 4) | θ value | 4.11246e-53 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13727, Q14227 | Gene names | PRG2, MBP | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (EMBP) (Pregnancy-associated major basic protein) (Proteoglycan 2, bone marrow) (BMPG). | |||||
|
REG1B_HUMAN
|
||||||
| NC score | 0.512165 (rank : 5) | θ value | 5.26297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
LIT1_MOUSE
|
||||||
| NC score | 0.506771 (rank : 6) | θ value | 2.44474e-05 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
LIT2_MOUSE
|
||||||
| NC score | 0.506115 (rank : 7) | θ value | 1.43324e-05 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
REG3G_MOUSE
|
||||||
| NC score | 0.500865 (rank : 8) | θ value | 4.1701e-05 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.489405 (rank : 9) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
REG3A_MOUSE
|
||||||
| NC score | 0.485866 (rank : 10) | θ value | 5.44631e-05 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.481569 (rank : 11) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
REG1A_HUMAN
|
||||||
| NC score | 0.475384 (rank : 12) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.469878 (rank : 13) | θ value | 1.69304e-06 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
REG3A_HUMAN
|
||||||
| NC score | 0.468637 (rank : 14) | θ value | 3.19293e-05 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.465657 (rank : 15) | θ value | 6.43352e-06 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.453129 (rank : 16) | θ value | 9.29e-05 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.452104 (rank : 17) | θ value | 2.44474e-05 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
REG3G_HUMAN
|
||||||
| NC score | 0.450250 (rank : 18) | θ value | 0.00020696 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
REG3B_MOUSE
|
||||||
| NC score | 0.449951 (rank : 19) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
FCER2_HUMAN
|
||||||
| NC score | 0.412137 (rank : 20) | θ value | 0.00102713 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
FCER2_MOUSE
|
||||||
| NC score | 0.396678 (rank : 21) | θ value | 3.77169e-06 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
REG4_HUMAN
|
||||||
| NC score | 0.389220 (rank : 22) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
REG4_MOUSE
|
||||||
| NC score | 0.367796 (rank : 23) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
TETN_HUMAN
|
||||||
| NC score | 0.364578 (rank : 24) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
TETN_MOUSE
|
||||||
| NC score | 0.364094 (rank : 25) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
CLC4G_MOUSE
|
||||||
| NC score | 0.357959 (rank : 26) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.346277 (rank : 27) | θ value | 0.000461057 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.336495 (rank : 28) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.335925 (rank : 29) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.332914 (rank : 30) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.332284 (rank : 31) | θ value | 9.29e-05 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.327470 (rank : 32) | θ value | 0.0330416 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
ASGR1_HUMAN
|
||||||
| NC score | 0.326733 (rank : 33) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
CLC4G_HUMAN
|
||||||
| NC score | 0.326004 (rank : 34) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.318348 (rank : 35) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.316028 (rank : 36) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.315606 (rank : 37) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
ASGR1_MOUSE
|
||||||
| NC score | 0.314460 (rank : 38) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
CLC4K_HUMAN
|
||||||
| NC score | 0.313280 (rank : 39) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
C209E_MOUSE
|
||||||
| NC score | 0.312877 (rank : 40) | θ value | 0.00228821 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.311271 (rank : 41) | θ value | 0.00035302 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CLC4K_MOUSE
|
||||||
| NC score | 0.305535 (rank : 42) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.301563 (rank : 43) | θ value | 8.40245e-06 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.301170 (rank : 44) | θ value | 6.43352e-06 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.300777 (rank : 45) | θ value | 0.000461057 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
ASGR2_HUMAN
|
||||||
| NC score | 0.299783 (rank : 46) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
CLC3A_MOUSE
|
||||||
| NC score | 0.297648 (rank : 47) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC3A_HUMAN
|
||||||
| NC score | 0.296957 (rank : 48) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
SFTA1_HUMAN
|
||||||
| NC score | 0.290034 (rank : 49) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
SFTPA_MOUSE
|
||||||
| NC score | 0.288557 (rank : 50) | θ value | 0.000461057 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35242 | Gene names | Sftpa1, Sftp-1, Sftp1, Sftpa | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein A precursor (SP-A) (PSP-A) (PSAP). | |||||
|
SFTA2_HUMAN
|
||||||
| NC score | 0.286887 (rank : 51) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
C209C_MOUSE
|
||||||
| NC score | 0.286192 (rank : 52) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
CLC4D_HUMAN
|
||||||
| NC score | 0.277989 (rank : 53) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
C209A_MOUSE
|
||||||
| NC score | 0.277211 (rank : 54) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
C209D_MOUSE
|
||||||
| NC score | 0.277086 (rank : 55) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
CLC4D_MOUSE
|
||||||
| NC score | 0.268223 (rank : 56) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
C209B_MOUSE
|
||||||
| NC score | 0.264721 (rank : 57) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
MBL1_MOUSE
|
||||||
| NC score | 0.263767 (rank : 58) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
CD209_HUMAN
|
||||||
| NC score | 0.262607 (rank : 59) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
CLC4E_MOUSE
|
||||||
| NC score | 0.260093 (rank : 60) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
SFTPD_MOUSE
|
||||||
| NC score | 0.256465 (rank : 61) | θ value | 4.1701e-05 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P50404 | Gene names | Sftpd, Sftp4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
CLC4M_HUMAN
|
||||||
| NC score | 0.256284 (rank : 62) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
MBL2_MOUSE
|
||||||
| NC score | 0.255859 (rank : 63) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
CLC4A_HUMAN
|
||||||
| NC score | 0.255375 (rank : 64) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
CLC6A_HUMAN
|
||||||
| NC score | 0.251697 (rank : 65) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
MBL2_HUMAN
|
||||||
| NC score | 0.250540 (rank : 66) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
CLC4E_HUMAN
|
||||||
| NC score | 0.250208 (rank : 67) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
SFTPD_HUMAN
|
||||||
| NC score | 0.248405 (rank : 68) | θ value | 0.00509761 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35247, Q86YK9, Q9UCJ2, Q9UCJ3 | Gene names | SFTPD, PSPD, SFTP4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
FREM1_MOUSE
|
||||||
| NC score | 0.245646 (rank : 69) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
CLC6A_MOUSE
|
||||||
| NC score | 0.242136 (rank : 70) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
KLRF1_HUMAN
|
||||||
| NC score | 0.241561 (rank : 71) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
CLC4A_MOUSE
|
||||||
| NC score | 0.241167 (rank : 72) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
NKG2D_MOUSE
|
||||||
| NC score | 0.234381 (rank : 73) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.233523 (rank : 74) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.232387 (rank : 75) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CLC4C_HUMAN
|
||||||
| NC score | 0.230811 (rank : 76) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
NKG2D_HUMAN
|
||||||
| NC score | 0.230101 (rank : 77) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
FREM1_HUMAN
|
||||||
| NC score | 0.222317 (rank : 78) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.214939 (rank : 79) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
KLRD1_HUMAN
|
||||||
| NC score | 0.213243 (rank : 80) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
CD69_HUMAN
|
||||||
| NC score | 0.206595 (rank : 81) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |

|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CLC11_MOUSE
|
||||||
| NC score | 0.201447 (rank : 82) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88200, Q8C946, Q9CTF0 | Gene names | Clec11a, Scgf | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin). | |||||
|
CD69_MOUSE
|
||||||
| NC score | 0.194171 (rank : 83) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
CLC11_HUMAN
|
||||||
| NC score | 0.187062 (rank : 84) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
CLC1A_MOUSE
|
||||||
| NC score | 0.183553 (rank : 85) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BWY2, Q7TPX7, Q8BMH5 | Gene names | Clec1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A. | |||||
|
KLRBB_MOUSE
|
||||||
| NC score | 0.181150 (rank : 86) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27812 | Gene names | Klrb1b, Ly55-b, Ly55b | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-34 (NKR-P1 34). | |||||
|
CLC9A_MOUSE
|
||||||
| NC score | 0.175436 (rank : 87) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRU4 | Gene names | Clec9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
NKG2E_HUMAN
|
||||||
| NC score | 0.173896 (rank : 88) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07444, Q96RL0, Q9UP04 | Gene names | KLRC3, NKG2E | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-E type II integral membrane protein (NKG2-E-activating NK receptor) (NK cell receptor E). | |||||
|
CLC9A_HUMAN
|
||||||
| NC score | 0.170126 (rank : 89) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UXN8 | Gene names | CLEC9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
NKG2C_HUMAN
|
||||||
| NC score | 0.169864 (rank : 90) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26717, O43802, Q9NR42 | Gene names | KLRC2, NKG2C | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-C type II integral membrane protein (NKG2-C-activating NK receptor) (NK cell receptor C) (CD159c antigen). | |||||
|
NKG2A_HUMAN
|
||||||
| NC score | 0.168341 (rank : 91) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P26715 | Gene names | KLRC1, NKG2A | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-A/NKG2-B type II integral membrane protein (NKG2-A/B-activating NK receptor) (NK cell receptor A) (CD159a antigen). | |||||
|
CLC2B_HUMAN
|
||||||
| NC score | 0.165254 (rank : 92) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92478, Q8IZE9, Q9BS74, Q9UQB4 | Gene names | CLEC2B, AICL, CLECSF2, IFNRG1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 2 member B (C-type lectin superfamily member 2) (Activation-induced C-type lectin) (IFN-alpha2b-inducing- related protein 1) (C-type lectin domain family 2 member B). | |||||
|
CLC5A_MOUSE
|
||||||
| NC score | 0.160860 (rank : 93) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R007, Q3UV97, Q8BL24 | Gene names | Clec5a, Clecsf5, Mdl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 5 member A (C-type lectin superfamily member 5) (Myeloid DAP12-associating lectin protein) (MDL-1). | |||||
|
KLRBA_MOUSE
|
||||||
| NC score | 0.160404 (rank : 94) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27811 | Gene names | Klrb1a, Ly55, Ly55a | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-2 (NKR-P1 2) (NKR-P1.7). | |||||
|
KLRBC_MOUSE
|
||||||
| NC score | 0.156957 (rank : 95) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27814 | Gene names | Klrb1c, Ly55-c, Ly55c | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-40 (NKR-P1 40) (NKR-P1.9). | |||||
|
LYAM1_MOUSE
|
||||||
| NC score | 0.152091 (rank : 96) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CLC1A_HUMAN
|
||||||
| NC score | 0.150772 (rank : 97) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NC01, Q8IUW7, Q9NZH3 | Gene names | CLEC1A, CLEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A (C-type lectin-like receptor 1) (CLEC-1). | |||||
|
OLR1_HUMAN
|
||||||
| NC score | 0.148821 (rank : 98) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.147901 (rank : 99) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CD72_HUMAN
|
||||||
| NC score | 0.146816 (rank : 100) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P21854 | Gene names | CD72 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2). | |||||
|
CLC5A_HUMAN
|
||||||
| NC score | 0.143684 (rank : 101) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NY25, Q9UKQ0 | Gene names | CLEC5A, CLECSF5, MDL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 5 member A (C-type lectin superfamily member 5) (Myeloid DAP12-associating lectin) (MDL-1). | |||||
|
CLC4F_HUMAN
|
||||||
| NC score | 0.136713 (rank : 102) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N1N0 | Gene names | CLEC4F, CLECSF13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13). | |||||
|
HPLN3_MOUSE
|
||||||
| NC score | 0.133782 (rank : 103) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
LYAM2_MOUSE
|
||||||
| NC score | 0.133713 (rank : 104) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
HPLN3_HUMAN
|
||||||
| NC score | 0.132361 (rank : 105) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
HPLN1_HUMAN
|
||||||
| NC score | 0.131014 (rank : 106) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| NC score | 0.130419 (rank : 107) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN2_MOUSE
|
||||||
| NC score | 0.130094 (rank : 108) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_HUMAN
|
||||||
| NC score | 0.129711 (rank : 109) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN4_HUMAN
|
||||||
| NC score | 0.128852 (rank : 110) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.127395 (rank : 111) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
OLR1_MOUSE
|
||||||
| NC score | 0.126361 (rank : 112) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
KLRA7_MOUSE
|
||||||
| NC score | 0.119393 (rank : 113) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60654, Q60655, Q60656, Q60683 | Gene names | Klra7, Ly-49g, Ly49-g, Ly49g, Ly49g4 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 7 (T-cell surface glycoprotein Ly- 49G) (Ly49-G antigen) (Lymphocyte antigen 49G). | |||||
|
LYAM2_HUMAN
|
||||||
| NC score | 0.118790 (rank : 114) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
KLRA3_MOUSE
|
||||||
| NC score | 0.116057 (rank : 115) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64329, Q61154, Q64257 | Gene names | Klra3, Ly-49c, Ly49C | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 3 (T-cell surface glycoprotein Ly- 49C) (Ly49-C antigen) (Lymphocyte antigen 49C) (5E6). | |||||
|
LY49A_MOUSE
|
||||||
| NC score | 0.113730 (rank : 116) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20937 | Gene names | Klra1, Ly-49, Ly-49a, Ly49, Ly49A | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein YE1/48 (T lymphocyte antigen A1) (Ly49-A antigen) (Lymphocyte antigen 49A). | |||||
|
KLRA5_MOUSE
|
||||||
| NC score | 0.111209 (rank : 117) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60652 | Gene names | Klra5, Ly-49e, Ly49-e, Ly49e | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 5 (T-cell surface glycoprotein Ly- 49E) (Ly49-E antigen) (Lymphocyte antigen 49E). | |||||
|
KLRA4_MOUSE
|
||||||
| NC score | 0.108446 (rank : 118) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60651, O78026, Q9EPA5 | Gene names | Klra4, Ly-49d, Ly49-d, Ly49d | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 4 (T-cell surface glycoprotein Ly- 49D) (Ly49-D antigen) (Lymphocyte antigen 49D). | |||||
|
KLRA6_MOUSE
|
||||||
| NC score | 0.101191 (rank : 119) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60653 | Gene names | Klra6, Ly-49f, Ly49-f, Ly49F | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 6 (T-cell surface glycoprotein Ly- 49F) (Ly49-F antigen) (Lymphocyte antigen 49F). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.100748 (rank : 120) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
LYAM3_HUMAN
|
||||||
| NC score | 0.100436 (rank : 121) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
KLRA8_MOUSE
|
||||||
| NC score | 0.097529 (rank : 122) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60682, O78027 | Gene names | Klra8, Ly-49h, Ly49-h, Ly49H | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 8 (T-cell surface glycoprotein Ly- 49H) (Ly49-H antigen) (Lymphocyte antigen 49H). | |||||
|
LYAM3_MOUSE
|
||||||
| NC score | 0.094623 (rank : 123) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.087439 (rank : 124) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.086348 (rank : 125) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.078156 (rank : 126) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.073765 (rank : 127) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
CD72_MOUSE
|
||||||
| NC score | 0.064319 (rank : 128) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21855, Q64225 | Gene names | Cd72, Ly-32, Lyb-2 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2) (Antigen Ly-32). | |||||
|
XLKD1_MOUSE
|
||||||
| NC score | 0.064185 (rank : 129) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_HUMAN
|
||||||
| NC score | 0.062544 (rank : 130) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.061564 (rank : 131) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.058895 (rank : 132) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.058726 (rank : 133) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.058248 (rank : 134) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.057546 (rank : 135) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TRBM_MOUSE
|
||||||
| NC score | 0.055911 (rank : 136) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.054599 (rank : 137) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.053635 (rank : 138) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.053619 (rank : 139) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.053099 (rank : 140) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.052523 (rank : 141) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.051228 (rank : 142) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.049240 (rank : 143) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.042115 (rank : 144) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.038646 (rank : 145) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.035858 (rank : 146) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.034570 (rank : 147) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
DPOE3_HUMAN
|
||||||
| NC score | 0.034542 (rank : 148) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.032902 (rank : 149) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.032509 (rank : 150) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.031422 (rank : 151) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.031151 (rank : 152) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.030071 (rank : 153) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.026154 (rank : 154) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.026013 (rank : 155) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
REN3B_HUMAN
|
||||||
| NC score | 0.023422 (rank : 156) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
D2HDH_MOUSE
|
||||||
| NC score | 0.022868 (rank : 157) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIM3, Q3TDF5, Q8BU06 | Gene names | D2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.022682 (rank : 158) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.022436 (rank : 159) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.021613 (rank : 160) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.019443 (rank : 161) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.019292 (rank : 162) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.018060 (rank : 163) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.017363 (rank : 164) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.016959 (rank : 165) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.015930 (rank : 166) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.015416 (rank : 167) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.014540 (rank : 168) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.013110 (rank : 169) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.010280 (rank : 170) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.010164 (rank : 171) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.009999 (rank : 172) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.009774 (rank : 173) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.008012 (rank : 174) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.007914 (rank : 175) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
AN13A_MOUSE
|
||||||
| NC score | 0.007816 (rank : 176) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UP5, Q2VPQ7, Q6P7F2 | Gene names | Ankrd13a, Ankrd13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13A. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.006953 (rank : 177) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.006679 (rank : 178) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.006466 (rank : 179) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.006309 (rank : 180) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.005259 (rank : 181) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.005162 (rank : 182) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.005037 (rank : 183) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZN646_HUMAN
|
||||||
| NC score | 0.004734 (rank : 184) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.004215 (rank : 185) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
FHL5_HUMAN
|
||||||
| NC score | 0.001907 (rank : 186) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | -0.002037 (rank : 187) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.002417 (rank : 188) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | -0.002633 (rank : 189) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||