Please be patient as the page loads
|
D2HDH_MOUSE
|
||||||
| SwissProt Accessions | Q8CIM3, Q3TDF5, Q8BU06 | Gene names | D2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
D2HDH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994663 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N465, Q6IQ24, Q8N5Q8 | Gene names | D2HGDH, D2HGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
D2HDH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CIM3, Q3TDF5, Q8BU06 | Gene names | D2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
ADAS_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 3) | NC score | 0.515910 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00116, Q2TU35 | Gene names | AGPS | |||
|
Domain Architecture |

|
|||||
| Description | Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor (EC 2.5.1.26) (Alkyl-DHAP synthase) (Alkylglycerone-phosphate synthase) (Aging-associated protein 5). | |||||
|
ADAS_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 4) | NC score | 0.512033 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0I1 | Gene names | Agps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor (EC 2.5.1.26) (Alkyl-DHAP synthase) (Alkylglycerone-phosphate synthase). | |||||
|
GGLO_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 5) | NC score | 0.461467 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58710 | Gene names | Gulo | |||
|
Domain Architecture |

|
|||||
| Description | L-gulonolactone oxidase (EC 1.1.3.8) (LGO) (L-gulono-gamma-lactone oxidase) (GLO). | |||||
|
DHC24_HUMAN
|
||||||
| θ value | 0.62314 (rank : 6) | NC score | 0.108790 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |
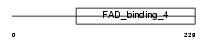
|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
MARK3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.009908 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.010253 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
K0020_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.026838 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15397, Q547G7, Q5SZY9, Q6IB47, Q96B27, Q96L78, Q96L79, Q96L80 | Gene names | KIAA0020, XTP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020 (HBV X-transactivated gene 5 protein) (Minor histocompatibility antigen HA-8) (HLA-HA8). | |||||
|
PRG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.022868 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.012893 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.013163 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
EI2BD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.017138 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
D2HDH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CIM3, Q3TDF5, Q8BU06 | Gene names | D2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
D2HDH_HUMAN
|
||||||
| NC score | 0.994663 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N465, Q6IQ24, Q8N5Q8 | Gene names | D2HGDH, D2HGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.-). | |||||
|
ADAS_HUMAN
|
||||||
| NC score | 0.515910 (rank : 3) | θ value | 3.76295e-14 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00116, Q2TU35 | Gene names | AGPS | |||
|
Domain Architecture |

|
|||||
| Description | Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor (EC 2.5.1.26) (Alkyl-DHAP synthase) (Alkylglycerone-phosphate synthase) (Aging-associated protein 5). | |||||
|
ADAS_MOUSE
|
||||||
| NC score | 0.512033 (rank : 4) | θ value | 1.42992e-13 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0I1 | Gene names | Agps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor (EC 2.5.1.26) (Alkyl-DHAP synthase) (Alkylglycerone-phosphate synthase). | |||||
|
GGLO_MOUSE
|
||||||
| NC score | 0.461467 (rank : 5) | θ value | 4.0297e-08 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58710 | Gene names | Gulo | |||
|
Domain Architecture |

|
|||||
| Description | L-gulonolactone oxidase (EC 1.1.3.8) (LGO) (L-gulono-gamma-lactone oxidase) (GLO). | |||||
|
DHC24_HUMAN
|
||||||
| NC score | 0.108790 (rank : 6) | θ value | 0.62314 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |

|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
K0020_HUMAN
|
||||||
| NC score | 0.026838 (rank : 7) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15397, Q547G7, Q5SZY9, Q6IB47, Q96B27, Q96L78, Q96L79, Q96L80 | Gene names | KIAA0020, XTP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020 (HBV X-transactivated gene 5 protein) (Minor histocompatibility antigen HA-8) (HLA-HA8). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.022868 (rank : 8) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
EI2BD_MOUSE
|
||||||
| NC score | 0.017138 (rank : 9) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.013163 (rank : 10) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.012893 (rank : 11) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | 0.010253 (rank : 12) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
MARK3_MOUSE
|
||||||
| NC score | 0.009908 (rank : 13) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||