Please be patient as the page loads
|
RUNX2_MOUSE
|
||||||
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RUNX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996222 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 1.20424e-145 (rank : 3) | NC score | 0.959613 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
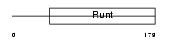
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 2.511e-143 (rank : 4) | NC score | 0.956527 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX3_HUMAN
|
||||||
| θ value | 1.21547e-113 (rank : 5) | NC score | 0.945060 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |
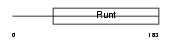
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 3.54468e-105 (rank : 6) | NC score | 0.938492 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
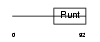
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.082771 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.087299 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.077697 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.035028 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.080911 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.030824 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.044596 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.056137 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.045333 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.032926 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.049696 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.038169 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.015767 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.045606 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.024898 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.062879 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.075694 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.059758 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
M3K9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.008128 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.038640 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.050233 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.017028 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.032473 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.053313 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.036490 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.009181 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.046489 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.037835 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.008723 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.012101 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.018769 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.030990 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.035179 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.011688 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.082133 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.032028 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FBX42_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.036661 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.014876 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.058333 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.053020 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.037444 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.036786 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.053138 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.032394 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.043865 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.026838 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
FBX42_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.035012 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDJ6, Q8CE76, Q99KY2 | Gene names | Fbxo42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.012401 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
MICA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.012026 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.023241 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.033397 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.007431 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.031732 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.017376 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.028325 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.032758 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.031058 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SF01_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.046685 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.044221 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.064585 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.021814 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
CD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.033631 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08920, Q61394 | Gene names | Cd2, Ly-37 | |||
|
Domain Architecture |
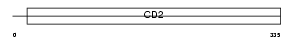
|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Lymphocyte antigen Ly-37). | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.007499 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.005242 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.025019 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.002550 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.005731 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.038168 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.063404 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TNFC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.029191 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41155 | Gene names | Ltb, Tnfc, Tnfsf3 | |||
|
Domain Architecture |
|
|||||
| Description | Lymphotoxin-beta (LT-beta) (Tumor necrosis factor C) (TNF-C) (Tumor necrosis factor ligand superfamily member 3). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.046373 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.031525 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.033097 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.026332 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.011315 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.020269 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.020379 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.021898 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.039258 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.020184 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.015702 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.019234 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.042583 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.001098 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.010660 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.013882 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.010692 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.009841 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.046577 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.013984 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.019320 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.018474 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.011061 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.016814 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ZMYM6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.010210 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.004982 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.001646 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.016506 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.003494 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
GAH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.010775 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013249 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.009731 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.016643 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.032225 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.003993 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.039630 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.021757 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.013154 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.009979 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.020562 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.001081 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.012827 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TSSC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.013270 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHE7 | Gene names | Tssc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.053675 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.072377 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.071288 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ZN645_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.057178 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.996222 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.959613 (rank : 3) | θ value | 1.20424e-145 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.956527 (rank : 4) | θ value | 2.511e-143 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX3_HUMAN
|
||||||
| NC score | 0.945060 (rank : 5) | θ value | 1.21547e-113 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.938492 (rank : 6) | θ value | 3.54468e-105 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.087299 (rank : 7) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.082771 (rank : 8) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.082133 (rank : 9) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.080911 (rank : 10) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.077697 (rank : 11) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.075694 (rank : 12) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.072377 (rank : 13) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.071288 (rank : 14) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.064585 (rank : 15) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.063404 (rank : 16) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.062879 (rank : 17) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.059758 (rank : 18) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.058333 (rank : 19) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.057178 (rank : 20) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.056137 (rank : 21) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.053675 (rank : 22) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.053313 (rank : 23) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.053138 (rank : 24) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.053020 (rank : 25) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.050233 (rank : 26) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.049696 (rank : 27) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.046685 (rank : 28) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.046577 (rank : 29) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.046489 (rank : 30) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.046373 (rank : 31) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.045606 (rank : 32) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.045333 (rank : 33) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.044596 (rank : 34) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.044221 (rank : 35) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.043865 (rank : 36) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.042583 (rank : 37) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.039630 (rank : 38) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.039258 (rank : 39) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.038640 (rank : 40) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.038169 (rank : 41) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.038168 (rank : 42) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.037835 (rank : 43) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.037444 (rank : 44) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.036786 (rank : 45) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
FBX42_HUMAN
|
||||||
| NC score | 0.036661 (rank : 46) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.036490 (rank : 47) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.035179 (rank : 48) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.035028 (rank : 49) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
FBX42_MOUSE
|
||||||
| NC score | 0.035012 (rank : 50) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDJ6, Q8CE76, Q99KY2 | Gene names | Fbxo42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
CD2_MOUSE
|
||||||
| NC score | 0.033631 (rank : 51) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08920, Q61394 | Gene names | Cd2, Ly-37 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Lymphocyte antigen Ly-37). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.033397 (rank : 52) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.033097 (rank : 53) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.032926 (rank : 54) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.032758 (rank : 55) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.032473 (rank : 56) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.032394 (rank : 57) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.032225 (rank : 58) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.032028 (rank : 59) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.031732 (rank : 60) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.031525 (rank : 61) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.031058 (rank : 62) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.030990 (rank : 63) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.030824 (rank : 64) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
TNFC_MOUSE
|
||||||
| NC score | 0.029191 (rank : 65) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41155 | Gene names | Ltb, Tnfc, Tnfsf3 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphotoxin-beta (LT-beta) (Tumor necrosis factor C) (TNF-C) (Tumor necrosis factor ligand superfamily member 3). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.028325 (rank : 66) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.026838 (rank : 67) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.026332 (rank : 68) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.025019 (rank : 69) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.024898 (rank : 70) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.023241 (rank : 71) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.021898 (rank : 72) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.021814 (rank : 73) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.021757 (rank : 74) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.020562 (rank : 75) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.020379 (rank : 76) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.020269 (rank : 77) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.020184 (rank : 78) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.019320 (rank : 79) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.019234 (rank : 80) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.018769 (rank : 81) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.018474 (rank : 82) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.017376 (rank : 83) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.017028 (rank : 84) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.016814 (rank : 85) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.016643 (rank : 86) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.016506 (rank : 87) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.015767 (rank : 88) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.015702 (rank : 89) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.014876 (rank : 90) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.013984 (rank : 91) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.013882 (rank : 92) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
TSSC4_MOUSE
|
||||||
| NC score | 0.013270 (rank : 93) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHE7 | Gene names | Tssc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4. | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.013249 (rank : 94) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.013154 (rank : 95) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.012827 (rank : 96) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.012401 (rank : 97) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.012101 (rank : 98) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.012026 (rank : 99) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.011688 (rank : 100) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.011315 (rank : 101) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.011061 (rank : 102) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
GAH6_HUMAN
|
||||||
| NC score | 0.010775 (rank : 103) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.010692 (rank : 104) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.010660 (rank : 105) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
ZMYM6_HUMAN
|
||||||
| NC score | 0.010210 (rank : 106) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.009979 (rank : 107) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.009841 (rank : 108) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.009731 (rank : 109) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.009181 (rank : 110) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.008723 (rank : 111) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
M3K9_HUMAN
|
||||||
| NC score | 0.008128 (rank : 112) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.007499 (rank : 113) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.007431 (rank : 114) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.005731 (rank : 115) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.005242 (rank : 116) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.004982 (rank : 117) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.003993 (rank : 118) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.003494 (rank : 119) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.002550 (rank : 120) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.001646 (rank : 121) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
FES_HUMAN
|
||||||
| NC score | 0.001098 (rank : 122) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.001081 (rank : 123) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||