Please be patient as the page loads
|
RUNX1_MOUSE
|
||||||
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
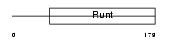
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RUNX1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985454 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 1.68038e-147 (rank : 3) | NC score | 0.964163 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 1.20424e-145 (rank : 4) | NC score | 0.959613 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 1.33764e-128 (rank : 5) | NC score | 0.955203 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
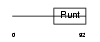
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_HUMAN
|
||||||
| θ value | 1.47893e-127 (rank : 6) | NC score | 0.960016 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |
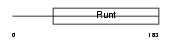
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 7) | NC score | 0.097695 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.061389 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.064549 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.041745 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.036434 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.069998 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.074242 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.044868 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.073263 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.064126 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.061677 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.022565 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
UBL7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.065044 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |
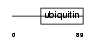
|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBL7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.065651 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.018117 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.019684 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PCDH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.005660 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.064445 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.017661 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.056683 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.053002 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.051385 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.048734 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.026227 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
TMM28_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.041043 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.016163 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.033204 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
SF01_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.050227 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.015739 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.068058 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.066503 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.025214 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.031586 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TBX15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.016206 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.030298 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
VAX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.015036 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
VAX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.013211 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.019535 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CDSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.036034 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.055096 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.024541 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.026035 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.004709 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.017370 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.004824 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
MARK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.004368 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.016401 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.041139 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.033005 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.028459 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.006619 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
P53_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.020543 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.008175 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.026876 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
RL12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.041637 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30050 | Gene names | RPL12 | |||
|
Domain Architecture |
|
|||||
| Description | 60S ribosomal protein L12. | |||||
|
RL12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.041637 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35979, Q9CQK4 | Gene names | Rpl12 | |||
|
Domain Architecture |
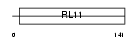
|
|||||
| Description | 60S ribosomal protein L12. | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.026653 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.005814 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.014535 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.008902 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.007645 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
NDF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.007414 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |
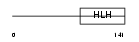
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.025661 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.012310 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SUOX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.021549 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51687 | Gene names | SUOX | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
APS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.007630 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.054899 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CRF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.016665 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
DMRT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.011650 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5R6, Q8IW77 | Gene names | DMRT1, DMT1 | |||
|
Domain Architecture |
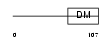
|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1 (DM domain expressed in testis protein 1). | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.007351 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
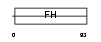
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
M3K9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.004368 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.005325 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.029785 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.024893 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.024838 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.031030 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.009800 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006007 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016361 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
AMPH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.016656 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.033346 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.005733 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | -0.000268 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.019126 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.020799 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.018954 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.009113 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.051404 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.985454 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.964163 (rank : 3) | θ value | 1.68038e-147 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX3_HUMAN
|
||||||
| NC score | 0.960016 (rank : 4) | θ value | 1.47893e-127 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.959613 (rank : 5) | θ value | 1.20424e-145 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.955203 (rank : 6) | θ value | 1.33764e-128 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.097695 (rank : 7) | θ value | 0.00175202 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.074242 (rank : 8) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.073263 (rank : 9) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.069998 (rank : 10) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.068058 (rank : 11) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.066503 (rank : 12) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
UBL7_MOUSE
|
||||||
| NC score | 0.065651 (rank : 13) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
UBL7_HUMAN
|
||||||
| NC score | 0.065044 (rank : 14) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.064549 (rank : 15) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.064445 (rank : 16) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.064126 (rank : 17) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.061677 (rank : 18) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.061389 (rank : 19) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.056683 (rank : 20) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.055096 (rank : 21) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.054899 (rank : 22) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.053002 (rank : 23) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.051404 (rank : 24) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.051385 (rank : 25) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.050227 (rank : 26) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.048734 (rank : 27) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.044868 (rank : 28) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.041745 (rank : 29) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
RL12_HUMAN
|
||||||
| NC score | 0.041637 (rank : 30) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30050 | Gene names | RPL12 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L12. | |||||
|
RL12_MOUSE
|
||||||
| NC score | 0.041637 (rank : 31) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35979, Q9CQK4 | Gene names | Rpl12 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L12. | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.041139 (rank : 32) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
TMM28_HUMAN
|
||||||
| NC score | 0.041043 (rank : 33) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.036434 (rank : 34) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.036034 (rank : 35) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.033346 (rank : 36) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.033204 (rank : 37) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.033005 (rank : 38) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.031586 (rank : 39) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.031030 (rank : 40) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.030298 (rank : 41) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.029785 (rank : 42) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.028459 (rank : 43) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.026876 (rank : 44) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.026653 (rank : 45) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.026227 (rank : 46) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.026035 (rank : 47) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.025661 (rank : 48) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.025214 (rank : 49) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.024893 (rank : 50) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.024838 (rank : 51) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.024541 (rank : 52) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.022565 (rank : 53) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
SUOX_HUMAN
|
||||||
| NC score | 0.021549 (rank : 54) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51687 | Gene names | SUOX | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.020799 (rank : 55) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.020543 (rank : 56) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.019684 (rank : 57) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.019535 (rank : 58) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.019126 (rank : 59) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.018954 (rank : 60) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.018117 (rank : 61) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.017661 (rank : 62) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.017370 (rank : 63) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
CRF_HUMAN
|
||||||
| NC score | 0.016665 (rank : 64) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
AMPH_MOUSE
|
||||||
| NC score | 0.016656 (rank : 65) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.016401 (rank : 66) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.016361 (rank : 67) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
TBX15_HUMAN
|
||||||
| NC score | 0.016206 (rank : 68) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.016163 (rank : 69) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.015739 (rank : 70) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
VAX1_HUMAN
|
||||||
| NC score | 0.015036 (rank : 71) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.014535 (rank : 72) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
VAX1_MOUSE
|
||||||
| NC score | 0.013211 (rank : 73) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.012310 (rank : 74) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
DMRT1_HUMAN
|
||||||
| NC score | 0.011650 (rank : 75) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5R6, Q8IW77 | Gene names | DMRT1, DMT1 | |||
|
Domain Architecture |

|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1 (DM domain expressed in testis protein 1). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.009800 (rank : 76) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.009113 (rank : 77) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.008902 (rank : 78) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.008175 (rank : 79) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.007645 (rank : 80) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
APS_HUMAN
|
||||||
| NC score | 0.007630 (rank : 81) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
NDF4_MOUSE
|
||||||
| NC score | 0.007414 (rank : 82) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.007351 (rank : 83) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.006619 (rank : 84) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.006007 (rank : 85) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.005814 (rank : 86) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.005733 (rank : 87) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
PCDH1_HUMAN
|
||||||
| NC score | 0.005660 (rank : 88) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.005325 (rank : 89) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | 0.004824 (rank : 90) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.004709 (rank : 91) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
M3K9_HUMAN
|
||||||
| NC score | 0.004368 (rank : 92) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
MARK3_MOUSE
|
||||||
| NC score | 0.004368 (rank : 93) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | -0.000268 (rank : 94) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||