Please be patient as the page loads
|
ETV6_HUMAN
|
||||||
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ETV6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985194 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 3) | NC score | 0.942091 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 4) | NC score | 0.848087 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 5) | NC score | 0.845715 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 6) | NC score | 0.818915 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 7) | NC score | 0.813063 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 8) | NC score | 0.800573 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 9) | NC score | 0.823634 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 10) | NC score | 0.824662 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 11) | NC score | 0.840539 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 12) | NC score | 0.816569 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 13) | NC score | 0.836908 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 14) | NC score | 0.823034 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 15) | NC score | 0.812162 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 16) | NC score | 0.780679 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 17) | NC score | 0.780334 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 18) | NC score | 0.827669 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 19) | NC score | 0.799014 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 20) | NC score | 0.781983 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 21) | NC score | 0.782924 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 22) | NC score | 0.820359 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 23) | NC score | 0.819671 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 24) | NC score | 0.816083 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 25) | NC score | 0.799950 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 26) | NC score | 0.834691 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 27) | NC score | 0.832946 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 28) | NC score | 0.777889 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 29) | NC score | 0.762444 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 30) | NC score | 0.804088 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 31) | NC score | 0.805272 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 32) | NC score | 0.834871 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 33) | NC score | 0.821412 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 34) | NC score | 0.822284 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 35) | NC score | 0.833547 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 36) | NC score | 0.769201 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 37) | NC score | 0.789549 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 38) | NC score | 0.785096 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 39) | NC score | 0.784580 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 40) | NC score | 0.803598 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 41) | NC score | 0.808101 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 42) | NC score | 0.627389 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 43) | NC score | 0.647587 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 44) | NC score | 0.637532 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 45) | NC score | 0.643572 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 46) | NC score | 0.626436 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 47) | NC score | 0.614938 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 48) | NC score | 0.051630 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 49) | NC score | 0.045955 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.057314 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.025014 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.038770 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.028942 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
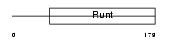
RUNX1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.029869 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.031004 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.039067 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.025223 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.014350 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.014013 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.046319 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.038220 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.035656 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.040643 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.024126 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.032861 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.048399 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.028000 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.014738 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.050710 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.041386 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.031006 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.022659 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PABP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.010619 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.010634 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.044100 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CK060_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.031162 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQC8 | Gene names | C11orf60, C11orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60. | |||||
|
CK060_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.031214 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DB07, Q91Z06, Q9JHT1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60 homolog. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.011908 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.024934 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
LSR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.018242 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
PSD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.016048 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
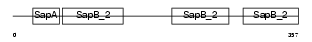
PSPB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.019802 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |
|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.024541 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.032669 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.014916 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.006873 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HSN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.035400 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
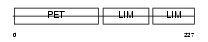
LMO6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.005220 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
PUS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.024807 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.010378 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.020593 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZN691_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | -0.004888 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV52, O95878, Q9NWE8 | Gene names | ZNF691 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 691. | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.005281 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
EPC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.010753 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
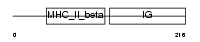
HB24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.004851 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.009513 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.032016 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.020788 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | -0.000609 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.014788 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.027744 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.011458 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
PJA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.008603 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.002401 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
SMBT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.010165 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.020290 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.005680 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.019828 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CE025_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.015245 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.003740 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.008224 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.002179 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
SMS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.010565 (rank : 103) | |||
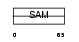
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.021509 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.017805 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.020689 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
HSN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.028541 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.003037 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.004213 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.015376 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.985194 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.942091 (rank : 3) | θ value | 9.48078e-50 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.848087 (rank : 4) | θ value | 1.63225e-17 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.845715 (rank : 5) | θ value | 1.63225e-17 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.840539 (rank : 6) | θ value | 2.35696e-16 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.836908 (rank : 7) | θ value | 3.07829e-16 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.834871 (rank : 8) | θ value | 3.76295e-14 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.834691 (rank : 9) | θ value | 2.20605e-14 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.833547 (rank : 10) | θ value | 1.86753e-13 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.832946 (rank : 11) | θ value | 2.20605e-14 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.827669 (rank : 12) | θ value | 5.8054e-15 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.824662 (rank : 13) | θ value | 4.74913e-17 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.823634 (rank : 14) | θ value | 4.74913e-17 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.823034 (rank : 15) | θ value | 3.40345e-15 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.822284 (rank : 16) | θ value | 1.09485e-13 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.821412 (rank : 17) | θ value | 1.09485e-13 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.820359 (rank : 18) | θ value | 1.29331e-14 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.819671 (rank : 19) | θ value | 1.29331e-14 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.818915 (rank : 20) | θ value | 2.7842e-17 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.816569 (rank : 21) | θ value | 3.07829e-16 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.816083 (rank : 22) | θ value | 2.20605e-14 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.813063 (rank : 23) | θ value | 2.7842e-17 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.812162 (rank : 24) | θ value | 3.40345e-15 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.808101 (rank : 25) | θ value | 2.98157e-11 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.805272 (rank : 26) | θ value | 2.88119e-14 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.804088 (rank : 27) | θ value | 2.88119e-14 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.803598 (rank : 28) | θ value | 2.98157e-11 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.800573 (rank : 29) | θ value | 3.63628e-17 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.799950 (rank : 30) | θ value | 2.20605e-14 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.799014 (rank : 31) | θ value | 5.8054e-15 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.789549 (rank : 32) | θ value | 2.43908e-13 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.785096 (rank : 33) | θ value | 3.52202e-12 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.784580 (rank : 34) | θ value | 3.52202e-12 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.782924 (rank : 35) | θ value | 5.8054e-15 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.781983 (rank : 36) | θ value | 5.8054e-15 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.780679 (rank : 37) | θ value | 4.44505e-15 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.780334 (rank : 38) | θ value | 4.44505e-15 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.777889 (rank : 39) | θ value | 2.88119e-14 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.769201 (rank : 40) | θ value | 2.43908e-13 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.762444 (rank : 41) | θ value | 2.88119e-14 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.647587 (rank : 42) | θ value | 1.17247e-07 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.643572 (rank : 43) | θ value | 2.44474e-05 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.637532 (rank : 44) | θ value | 2.44474e-05 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.627389 (rank : 45) | θ value | 6.87365e-08 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.626436 (rank : 46) | θ value | 5.44631e-05 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.614938 (rank : 47) | θ value | 0.000270298 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.057314 (rank : 48) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.051630 (rank : 49) | θ value | 0.00390308 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.050710 (rank : 50) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.048399 (rank : 51) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.046319 (rank : 52) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.045955 (rank : 53) | θ value | 0.0252991 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.044100 (rank : 54) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.041386 (rank : 55) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.040643 (rank : 56) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.039067 (rank : 57) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.038770 (rank : 58) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.038220 (rank : 59) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.035656 (rank : 60) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
HSN2_HUMAN
|
||||||
| NC score | 0.035400 (rank : 61) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.032861 (rank : 62) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.032669 (rank : 63) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.032016 (rank : 64) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CK060_MOUSE
|
||||||
| NC score | 0.031214 (rank : 65) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DB07, Q91Z06, Q9JHT1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60 homolog. | |||||
|
CK060_HUMAN
|
||||||
| NC score | 0.031162 (rank : 66) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQC8 | Gene names | C11orf60, C11orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60. | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.031006 (rank : 67) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.031004 (rank : 68) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.029869 (rank : 69) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.028942 (rank : 70) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
HSN2_MOUSE
|
||||||
| NC score | 0.028541 (rank : 71) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.028000 (rank : 72) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.027744 (rank : 73) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.025223 (rank : 74) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.025014 (rank : 75) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.024934 (rank : 76) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
PUS3_HUMAN
|
||||||
| NC score | 0.024807 (rank : 77) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.024541 (rank : 78) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.024126 (rank : 79) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.022659 (rank : 80) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.021509 (rank : 81) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.020788 (rank : 82) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.020689 (rank : 83) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.020593 (rank : 84) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.020290 (rank : 85) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.019828 (rank : 86) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
PSPB_HUMAN
|
||||||
| NC score | 0.019802 (rank : 87) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
LSR_HUMAN
|
||||||
| NC score | 0.018242 (rank : 88) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.017805 (rank : 89) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.016048 (rank : 90) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.015376 (rank : 91) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CE025_HUMAN
|
||||||
| NC score | 0.015245 (rank : 92) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.014916 (rank : 93) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.014788 (rank : 94) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.014738 (rank : 95) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.014350 (rank : 96) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.014013 (rank : 97) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.011908 (rank : 98) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.011458 (rank : 99) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
EPC2_HUMAN
|
||||||
| NC score | 0.010753 (rank : 100) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.010634 (rank : 101) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.010619 (rank : 102) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
SMS1_MOUSE
|
||||||
| NC score | 0.010565 (rank : 103) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.010378 (rank : 104) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SMBT1_MOUSE
|
||||||
| NC score | 0.010165 (rank : 105) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.009513 (rank : 106) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PJA1_HUMAN
|
||||||
| NC score | 0.008603 (rank : 107) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.008224 (rank : 108) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.006873 (rank : 109) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.005680 (rank : 110) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.005281 (rank : 111) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.005220 (rank : 112) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
HB24_HUMAN
|
||||||
| NC score | 0.004851 (rank : 113) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.004213 (rank : 114) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.003740 (rank : 115) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.002401 (rank : 116) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.002179 (rank : 117) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.000609 (rank : 118) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | -0.003037 (rank : 119) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
ZN691_HUMAN
|
||||||
| NC score | -0.004888 (rank : 120) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV52, O95878, Q9NWE8 | Gene names | ZNF691 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 691. | |||||