Please be patient as the page loads
|
ETV1_HUMAN
|
||||||
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ETV1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998475 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 1.19589e-169 (rank : 3) | NC score | 0.980633 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 2.58046e-164 (rank : 4) | NC score | 0.960327 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 1.74296e-136 (rank : 5) | NC score | 0.975419 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.29305e-135 (rank : 6) | NC score | 0.976116 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 7) | NC score | 0.876984 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 2.19584e-30 (rank : 8) | NC score | 0.877090 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 1.4233e-29 (rank : 9) | NC score | 0.874461 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 10) | NC score | 0.873845 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 11) | NC score | 0.877156 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 12) | NC score | 0.848785 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERG_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 13) | NC score | 0.848320 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 14) | NC score | 0.862539 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 15) | NC score | 0.850008 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 16) | NC score | 0.851507 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 17) | NC score | 0.877646 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 18) | NC score | 0.875799 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 19) | NC score | 0.877698 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 20) | NC score | 0.863221 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 21) | NC score | 0.861758 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 22) | NC score | 0.874800 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 23) | NC score | 0.880793 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 24) | NC score | 0.875478 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 25) | NC score | 0.819501 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 26) | NC score | 0.818723 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 27) | NC score | 0.787708 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 28) | NC score | 0.810524 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 29) | NC score | 0.766902 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 30) | NC score | 0.785513 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 31) | NC score | 0.779090 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 32) | NC score | 0.787010 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 33) | NC score | 0.787606 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 34) | NC score | 0.823961 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 35) | NC score | 0.822262 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 36) | NC score | 0.787013 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 37) | NC score | 0.781983 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 38) | NC score | 0.783574 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 39) | NC score | 0.805334 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 40) | NC score | 0.767817 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 41) | NC score | 0.767311 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 42) | NC score | 0.588879 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 43) | NC score | 0.580034 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 44) | NC score | 0.581165 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 45) | NC score | 0.571713 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.051152 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 47) | NC score | 0.036237 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 48) | NC score | 0.032152 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.044489 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
F113A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.045236 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.028713 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.029608 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.017849 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.028467 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.053205 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.018041 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.025233 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.028101 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.017942 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.032146 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.023223 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BCL7C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.029056 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.019553 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.011073 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.010831 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.027171 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.030276 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.020247 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
KLF5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | -0.000207 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
TAF9B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.014277 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.015847 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.007469 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.021409 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
DMWD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.005515 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.003373 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
LACTB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.010293 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP89 | Gene names | Lactb, Lact1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.014894 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.024159 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.007809 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.008875 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.008646 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | -0.001135 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.007923 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.006636 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.002352 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
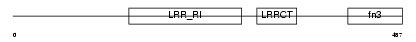
FLRT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | -0.000541 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43155 | Gene names | FLRT2, KIAA0405 | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT2 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 2). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.013792 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.010712 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.009602 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.006265 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.007419 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.036281 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.003000 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
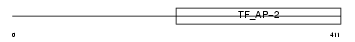
AP2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.008910 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05549, Q13777 | Gene names | TFAP2A, AP2TF, TFAP2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor AP-2 alpha (AP2-alpha) (Activating enhancer- binding protein 2 alpha) (AP-2 transcription factor) (Activator protein 2) (AP-2). | |||||
|
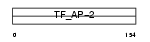
AP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.009066 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P34056, Q60740, Q60741, Q60742, Q60743, Q62067, Q62068, Q62069, Q91VX0, Q9CRY4 | Gene names | Tfap2a, Ap2tf, Tcfap2a | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor AP-2 alpha (AP2-alpha) (Activating enhancer- binding protein 2 alpha) (Activator protein 2) (AP-2). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.002079 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | -0.000468 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.004451 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.005063 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
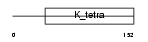
HIC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | -0.003411 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.003669 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
KLF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.000952 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||
|
PLAL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | -0.004766 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPG8, Q92584 | Gene names | PLAGL2, KIAA0198 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein PLAGL2 (Pleiomorphic adenoma-like protein 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | -0.002664 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.024465 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.032641 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.009116 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
SPIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.457895 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.434412 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.998475 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.980633 (rank : 3) | θ value | 1.19589e-169 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.976116 (rank : 4) | θ value | 4.29305e-135 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.975419 (rank : 5) | θ value | 1.74296e-136 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.960327 (rank : 6) | θ value | 2.58046e-164 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.880793 (rank : 7) | θ value | 1.5242e-23 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.877698 (rank : 8) | θ value | 1.05554e-24 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.877646 (rank : 9) | θ value | 7.30988e-26 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.877156 (rank : 10) | θ value | 2.27234e-27 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.877090 (rank : 11) | θ value | 2.19584e-30 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.876984 (rank : 12) | θ value | 2.19584e-30 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.875799 (rank : 13) | θ value | 7.30988e-26 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.875478 (rank : 14) | θ value | 2.20094e-22 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.874800 (rank : 15) | θ value | 3.07116e-24 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.874461 (rank : 16) | θ value | 1.4233e-29 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.873845 (rank : 17) | θ value | 1.4233e-29 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.863221 (rank : 18) | θ value | 1.05554e-24 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.862539 (rank : 19) | θ value | 1.47289e-26 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.861758 (rank : 20) | θ value | 2.35151e-24 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.851507 (rank : 21) | θ value | 4.28545e-26 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.850008 (rank : 22) | θ value | 4.28545e-26 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.848785 (rank : 23) | θ value | 5.06226e-27 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.848320 (rank : 24) | θ value | 8.63488e-27 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.823961 (rank : 25) | θ value | 3.07829e-16 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.822262 (rank : 26) | θ value | 4.02038e-16 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.819501 (rank : 27) | θ value | 2.97466e-19 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.818723 (rank : 28) | θ value | 2.97466e-19 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.810524 (rank : 29) | θ value | 3.28887e-18 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.805334 (rank : 30) | θ value | 1.74796e-11 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.787708 (rank : 31) | θ value | 3.28887e-18 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.787606 (rank : 32) | θ value | 2.35696e-16 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.787013 (rank : 33) | θ value | 1.52774e-15 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.787010 (rank : 34) | θ value | 6.20254e-17 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.785513 (rank : 35) | θ value | 6.20254e-17 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.783574 (rank : 36) | θ value | 6.41864e-14 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.781983 (rank : 37) | θ value | 5.8054e-15 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.779090 (rank : 38) | θ value | 6.20254e-17 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.767817 (rank : 39) | θ value | 2.98157e-11 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.767311 (rank : 40) | θ value | 1.47974e-10 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.766902 (rank : 41) | θ value | 4.74913e-17 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.588879 (rank : 42) | θ value | 4.0297e-08 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.581165 (rank : 43) | θ value | 3.77169e-06 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.580034 (rank : 44) | θ value | 3.77169e-06 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.571713 (rank : 45) | θ value | 3.77169e-06 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.457895 (rank : 46) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.434412 (rank : 47) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.053205 (rank : 48) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.051152 (rank : 49) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
F113A_MOUSE
|
||||||
| NC score | 0.045236 (rank : 50) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.044489 (rank : 51) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.036281 (rank : 52) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.036237 (rank : 53) | θ value | 0.0736092 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.032641 (rank : 54) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.032152 (rank : 55) | θ value | 0.0961366 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.032146 (rank : 56) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.030276 (rank : 57) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.029608 (rank : 58) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
BCL7C_HUMAN
|
||||||
| NC score | 0.029056 (rank : 59) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.028713 (rank : 60) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.028467 (rank : 61) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.028101 (rank : 62) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.027171 (rank : 63) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.025233 (rank : 64) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.024465 (rank : 65) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.024159 (rank : 66) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.023223 (rank : 67) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.021409 (rank : 68) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.020247 (rank : 69) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.019553 (rank : 70) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.018041 (rank : 71) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.017942 (rank : 72) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.017849 (rank : 73) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.015847 (rank : 74) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.014894 (rank : 75) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
TAF9B_HUMAN
|
||||||
| NC score | 0.014277 (rank : 76) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.013792 (rank : 77) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.011073 (rank : 78) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.010831 (rank : 79) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.010712 (rank : 80) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
LACTB_MOUSE
|
||||||
| NC score | 0.010293 (rank : 81) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP89 | Gene names | Lactb, Lact1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.009602 (rank : 82) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.009116 (rank : 83) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
AP2A_MOUSE
|
||||||
| NC score | 0.009066 (rank : 84) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P34056, Q60740, Q60741, Q60742, Q60743, Q62067, Q62068, Q62069, Q91VX0, Q9CRY4 | Gene names | Tfap2a, Ap2tf, Tcfap2a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 alpha (AP2-alpha) (Activating enhancer- binding protein 2 alpha) (Activator protein 2) (AP-2). | |||||
|
AP2A_HUMAN
|
||||||
| NC score | 0.008910 (rank : 85) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05549, Q13777 | Gene names | TFAP2A, AP2TF, TFAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 alpha (AP2-alpha) (Activating enhancer- binding protein 2 alpha) (AP-2 transcription factor) (Activator protein 2) (AP-2). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.008875 (rank : 86) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.008646 (rank : 87) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.007923 (rank : 88) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.007809 (rank : 89) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.007469 (rank : 90) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.007419 (rank : 91) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.006636 (rank : 92) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.006265 (rank : 93) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
DMWD_MOUSE
|
||||||
| NC score | 0.005515 (rank : 94) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.005063 (rank : 95) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.004451 (rank : 96) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.003669 (rank : 97) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.003373 (rank : 98) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.003000 (rank : 99) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.002352 (rank : 100) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.002079 (rank : 101) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
KLF5_HUMAN
|
||||||
| NC score | -0.000207 (rank : 102) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | -0.000468 (rank : 103) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
FLRT2_HUMAN
|
||||||
| NC score | -0.000541 (rank : 104) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43155 | Gene names | FLRT2, KIAA0405 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT2 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 2). | |||||
|
KLF5_MOUSE
|
||||||
| NC score | -0.000952 (rank : 105) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | -0.001135 (rank : 106) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.002664 (rank : 107) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
HIC1_MOUSE
|
||||||
| NC score | -0.003411 (rank : 108) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
PLAL2_HUMAN
|
||||||
| NC score | -0.004766 (rank : 109) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPG8, Q92584 | Gene names | PLAGL2, KIAA0198 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein PLAGL2 (Pleiomorphic adenoma-like protein 2). | |||||