Please be patient as the page loads
|
ETV6_MOUSE
|
||||||
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ETV6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985194 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 1.23823e-49 (rank : 3) | NC score | 0.944389 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 4) | NC score | 0.843693 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 5) | NC score | 0.843452 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 6) | NC score | 0.842713 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 7) | NC score | 0.851656 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 8) | NC score | 0.849211 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 9) | NC score | 0.819931 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 10) | NC score | 0.812640 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 11) | NC score | 0.825161 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 12) | NC score | 0.826225 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 13) | NC score | 0.844233 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 14) | NC score | 0.798704 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 15) | NC score | 0.840648 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 16) | NC score | 0.817645 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.832312 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 18) | NC score | 0.830214 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 19) | NC score | 0.803554 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 20) | NC score | 0.781739 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 21) | NC score | 0.781024 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 22) | NC score | 0.818083 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 23) | NC score | 0.804678 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 24) | NC score | 0.836426 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 25) | NC score | 0.834886 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 26) | NC score | 0.778423 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 27) | NC score | 0.763797 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 28) | NC score | 0.808942 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 29) | NC score | 0.809791 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 30) | NC score | 0.840865 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 31) | NC score | 0.784463 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 32) | NC score | 0.783574 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 33) | NC score | 0.820912 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 34) | NC score | 0.821867 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 35) | NC score | 0.839408 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 36) | NC score | 0.771750 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 37) | NC score | 0.793954 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 38) | NC score | 0.790422 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 39) | NC score | 0.789775 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 40) | NC score | 0.810634 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 41) | NC score | 0.815144 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 42) | NC score | 0.630063 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 43) | NC score | 0.650494 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 44) | NC score | 0.649402 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 45) | NC score | 0.643159 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 46) | NC score | 0.630437 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 47) | NC score | 0.619604 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 48) | NC score | 0.060435 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 0.125558 (rank : 49) | NC score | 0.041712 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.033044 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.026853 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.018158 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.016868 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.025746 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.018070 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.027308 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HSN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.043367 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.024702 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.020228 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TOB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.021485 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61471 | Gene names | Tob1, Tob, Trob | |||
|
Domain Architecture |

|
|||||
| Description | Tob1 protein (Transducer of erbB-2 1). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.027389 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.025667 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
HSN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.036755 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
SLIK5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.003215 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.015351 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.025894 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | -0.000862 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | -0.000826 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.015023 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PUS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.024833 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.010657 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.029404 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CALRL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.004895 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1W5, Q9QXH8, Q9WUP2 | Gene names | Calcrl, Crlr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.019381 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
HB24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.004929 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |
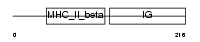
|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.021133 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.016748 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NRK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | -0.002315 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.020094 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SCMH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.017245 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.006044 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
UFO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | -0.002867 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00993 | Gene names | Axl, Ark, Ufo | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (Adhesion-related kinase). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.024787 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.012279 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PKN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | -0.002029 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70268, Q7TST2 | Gene names | Pkn1, Pkn, Prk1, Prkcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.021986 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.024134 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SCMH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.018939 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.022999 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.008733 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.000609 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.016430 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.030730 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PKN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | -0.002181 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16512, Q15143, Q8IUV5 | Gene names | PKN1, PKN, PRK1, PRKCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N) (Protein kinase PKN-alpha). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006676 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CYLD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.007723 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.007740 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
FOXGB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.001055 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
HDAC7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.012081 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
ITAL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.001904 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
SMS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009737 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |
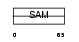
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SRTD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.008853 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.025928 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.985194 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.944389 (rank : 3) | θ value | 1.23823e-49 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.851656 (rank : 4) | θ value | 1.63225e-17 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.849211 (rank : 5) | θ value | 1.63225e-17 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.844233 (rank : 6) | θ value | 8.10077e-17 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.843693 (rank : 7) | θ value | 4.73814e-25 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.843452 (rank : 8) | θ value | 6.84181e-24 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.842713 (rank : 9) | θ value | 4.43474e-23 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.840865 (rank : 10) | θ value | 3.76295e-14 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.840648 (rank : 11) | θ value | 3.07829e-16 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.839408 (rank : 12) | θ value | 1.86753e-13 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.836426 (rank : 13) | θ value | 2.20605e-14 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.834886 (rank : 14) | θ value | 2.20605e-14 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.832312 (rank : 15) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.830214 (rank : 16) | θ value | 5.8054e-15 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.826225 (rank : 17) | θ value | 4.74913e-17 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.825161 (rank : 18) | θ value | 4.74913e-17 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.821867 (rank : 19) | θ value | 1.09485e-13 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.820912 (rank : 20) | θ value | 1.09485e-13 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.819931 (rank : 21) | θ value | 2.7842e-17 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.818083 (rank : 22) | θ value | 2.20605e-14 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.817645 (rank : 23) | θ value | 4.02038e-16 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.815144 (rank : 24) | θ value | 2.98157e-11 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.812640 (rank : 25) | θ value | 2.7842e-17 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.810634 (rank : 26) | θ value | 1.33837e-11 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.809791 (rank : 27) | θ value | 2.88119e-14 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.808942 (rank : 28) | θ value | 2.88119e-14 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.804678 (rank : 29) | θ value | 2.20605e-14 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.803554 (rank : 30) | θ value | 5.8054e-15 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.798704 (rank : 31) | θ value | 1.058e-16 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.793954 (rank : 32) | θ value | 2.43908e-13 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.790422 (rank : 33) | θ value | 3.52202e-12 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.789775 (rank : 34) | θ value | 3.52202e-12 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.784463 (rank : 35) | θ value | 4.91457e-14 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.783574 (rank : 36) | θ value | 6.41864e-14 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.781739 (rank : 37) | θ value | 1.68911e-14 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.781024 (rank : 38) | θ value | 1.68911e-14 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.778423 (rank : 39) | θ value | 2.88119e-14 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.771750 (rank : 40) | θ value | 2.43908e-13 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.763797 (rank : 41) | θ value | 2.88119e-14 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.650494 (rank : 42) | θ value | 8.97725e-08 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.649402 (rank : 43) | θ value | 9.92553e-07 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.643159 (rank : 44) | θ value | 2.44474e-05 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.630437 (rank : 45) | θ value | 0.000121331 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.630063 (rank : 46) | θ value | 6.87365e-08 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.619604 (rank : 47) | θ value | 0.000270298 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.060435 (rank : 48) | θ value | 0.0148317 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
HSN2_HUMAN
|
||||||
| NC score | 0.043367 (rank : 49) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.041712 (rank : 50) | θ value | 0.125558 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
HSN2_MOUSE
|
||||||
| NC score | 0.036755 (rank : 51) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.033044 (rank : 52) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.030730 (rank : 53) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.029404 (rank : 54) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.027389 (rank : 55) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.027308 (rank : 56) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.026853 (rank : 57) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.025928 (rank : 58) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.025894 (rank : 59) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.025746 (rank : 60) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.025667 (rank : 61) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
PUS3_HUMAN
|
||||||
| NC score | 0.024833 (rank : 62) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.024787 (rank : 63) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.024702 (rank : 64) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.024134 (rank : 65) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.022999 (rank : 66) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.021986 (rank : 67) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
TOB1_MOUSE
|
||||||
| NC score | 0.021485 (rank : 68) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61471 | Gene names | Tob1, Tob, Trob | |||
|
Domain Architecture |

|
|||||
| Description | Tob1 protein (Transducer of erbB-2 1). | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.021133 (rank : 69) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.020228 (rank : 70) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.020094 (rank : 71) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.019381 (rank : 72) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
SCMH1_MOUSE
|
||||||
| NC score | 0.018939 (rank : 73) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.018158 (rank : 74) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.018070 (rank : 75) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SCMH1_HUMAN
|
||||||
| NC score | 0.017245 (rank : 76) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.016868 (rank : 77) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.016748 (rank : 78) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.016430 (rank : 79) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.015351 (rank : 80) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.015023 (rank : 81) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.012279 (rank : 82) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
HDAC7_HUMAN
|
||||||
| NC score | 0.012081 (rank : 83) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.010657 (rank : 84) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
SMS1_MOUSE
|
||||||
| NC score | 0.009737 (rank : 85) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.008853 (rank : 86) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.008733 (rank : 87) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.007740 (rank : 88) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
CYLD_MOUSE
|
||||||
| NC score | 0.007723 (rank : 89) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.006676 (rank : 90) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.006044 (rank : 91) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HB24_HUMAN
|
||||||
| NC score | 0.004929 (rank : 92) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
CALRL_MOUSE
|
||||||
| NC score | 0.004895 (rank : 93) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1W5, Q9QXH8, Q9WUP2 | Gene names | Calcrl, Crlr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
SLIK5_MOUSE
|
||||||
| NC score | 0.003215 (rank : 94) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
ITAL_MOUSE
|
||||||
| NC score | 0.001904 (rank : 95) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
FOXGB_MOUSE
|
||||||
| NC score | 0.001055 (rank : 96) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.000609 (rank : 97) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | -0.000826 (rank : 98) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | -0.000862 (rank : 99) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PKN1_MOUSE
|
||||||
| NC score | -0.002029 (rank : 100) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70268, Q7TST2 | Gene names | Pkn1, Pkn, Prk1, Prkcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N). | |||||
|
PKN1_HUMAN
|
||||||
| NC score | -0.002181 (rank : 101) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16512, Q15143, Q8IUV5 | Gene names | PKN1, PKN, PRK1, PRKCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N) (Protein kinase PKN-alpha). | |||||
|
NRK_HUMAN
|
||||||
| NC score | -0.002315 (rank : 102) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
UFO_MOUSE
|
||||||
| NC score | -0.002867 (rank : 103) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00993 | Gene names | Axl, Ark, Ufo | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (Adhesion-related kinase). | |||||