Please be patient as the page loads
|
SCMH1_HUMAN
|
||||||
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SCMH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SCMH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992750 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SCML2_HUMAN
|
||||||
| θ value | 1.12714e-143 (rank : 3) | NC score | 0.965975 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQR0, Q86U98, Q8IWD0, Q8NDP2, Q9UGC5 | Gene names | SCML2 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 2. | |||||
|
LMBTL_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 4) | NC score | 0.782633 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
LMBL3_HUMAN
|
||||||
| θ value | 3.99248e-40 (rank : 5) | NC score | 0.826307 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 6) | NC score | 0.802391 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
LMBL2_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 7) | NC score | 0.766495 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59178 | Gene names | L3mbtl2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
LMBL2_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 8) | NC score | 0.764135 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969R5, Q8TEN1, Q96SC4, Q9BQI2, Q9UGS4 | Gene names | L3MBTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
SMBT1_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 9) | NC score | 0.722758 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
SMBT2_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 10) | NC score | 0.720073 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VUG0, Q9HCF5 | Gene names | SFMBT2, KIAA1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SCML1_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 11) | NC score | 0.728395 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UN30 | Gene names | SCML1 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 1. | |||||
|
SMBT1_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 12) | NC score | 0.713549 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
SMBT2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 13) | NC score | 0.713637 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 14) | NC score | 0.523110 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 15) | NC score | 0.521739 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 16) | NC score | 0.478474 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 17) | NC score | 0.514535 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 18) | NC score | 0.502267 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 19) | NC score | 0.504451 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 20) | NC score | 0.084500 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.036172 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.117548 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.113432 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.060305 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
MLTK_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.015198 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
CDAN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.060614 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWY9, Q7Z7L5, Q969N3 | Gene names | CDAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Codanin-1. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.014640 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.018217 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.024606 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.018865 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
LATS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.004946 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSJ6, Q8CDJ4, Q9JMI3 | Gene names | Lats2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.044927 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.032559 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
K0319_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.034344 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.008389 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.005004 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
VP13B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.042484 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.020196 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.027744 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.011655 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025534 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.059871 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.052096 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.023201 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
DRBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.011071 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.003395 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.003712 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CDC5L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.022918 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.017245 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
I12R2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.009694 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97378 | Gene names | Il12rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
K6PF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.009578 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08237, Q16814, Q16815 | Gene names | PFKM | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructokinase, muscle type (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme A) (PFK-A) (Phosphofructokinase-M). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.056053 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
P66B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.019927 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHR5, Q8C9Q3 | Gene names | Gatad2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 beta (p66/p68) (GATA zinc finger domain- containing protein 2B). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.003198 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.001473 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.012718 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MMAC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.024715 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4U1, Q5T157, Q9BRQ7 | Gene names | MMACHC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methylmalonic aciduria and homocystinuria type C protein. | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.003686 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.057765 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.017147 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.017575 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.005824 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CS018_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.021185 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEA5 | Gene names | C19orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf18 precursor. | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.024160 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
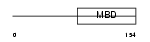
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.043302 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.018930 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.005096 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CDC5L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.019496 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.005597 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.015034 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.001420 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.017155 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.000961 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
RFTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.008840 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A0D4, Q3U654, Q3U8P6, Q80US1 | Gene names | Rftn1, Kiaa0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein). | |||||
|
SAM10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.050230 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.031802 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.006441 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.007634 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZN707_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.003271 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96C28 | Gene names | ZNF707 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 707. | |||||
|
SCMH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SCMH1_MOUSE
|
||||||
| NC score | 0.992750 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SCML2_HUMAN
|
||||||
| NC score | 0.965975 (rank : 3) | θ value | 1.12714e-143 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQR0, Q86U98, Q8IWD0, Q8NDP2, Q9UGC5 | Gene names | SCML2 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 2. | |||||
|
LMBL3_HUMAN
|
||||||
| NC score | 0.826307 (rank : 4) | θ value | 3.99248e-40 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.802391 (rank : 5) | θ value | 2.86122e-38 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
LMBTL_HUMAN
|
||||||
| NC score | 0.782633 (rank : 6) | θ value | 2.76489e-41 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
LMBL2_MOUSE
|
||||||
| NC score | 0.766495 (rank : 7) | θ value | 8.3442e-30 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59178 | Gene names | L3mbtl2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
LMBL2_HUMAN
|
||||||
| NC score | 0.764135 (rank : 8) | θ value | 4.14116e-29 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969R5, Q8TEN1, Q96SC4, Q9BQI2, Q9UGS4 | Gene names | L3MBTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
SCML1_HUMAN
|
||||||
| NC score | 0.728395 (rank : 9) | θ value | 1.02238e-19 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UN30 | Gene names | SCML1 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 1. | |||||
|
SMBT1_MOUSE
|
||||||
| NC score | 0.722758 (rank : 10) | θ value | 3.75424e-22 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
SMBT2_HUMAN
|
||||||
| NC score | 0.720073 (rank : 11) | θ value | 7.82807e-20 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VUG0, Q9HCF5 | Gene names | SFMBT2, KIAA1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SMBT2_MOUSE
|
||||||
| NC score | 0.713637 (rank : 12) | θ value | 1.24977e-17 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SMBT1_HUMAN
|
||||||
| NC score | 0.713549 (rank : 13) | θ value | 6.62687e-19 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.523110 (rank : 14) | θ value | 1.74796e-11 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.521739 (rank : 15) | θ value | 2.98157e-11 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.514535 (rank : 16) | θ value | 1.47974e-10 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.504451 (rank : 17) | θ value | 1.38499e-08 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.502267 (rank : 18) | θ value | 1.06045e-08 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.478474 (rank : 19) | θ value | 1.47974e-10 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.117548 (rank : 20) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.113432 (rank : 21) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.084500 (rank : 22) | θ value | 0.00228821 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CDAN1_HUMAN
|
||||||
| NC score | 0.060614 (rank : 23) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWY9, Q7Z7L5, Q969N3 | Gene names | CDAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Codanin-1. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.060305 (rank : 24) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.059871 (rank : 25) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.057765 (rank : 26) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.056053 (rank : 27) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.052096 (rank : 28) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SAM10_MOUSE
|
||||||
| NC score | 0.050230 (rank : 29) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.044927 (rank : 30) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.043302 (rank : 31) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
VP13B_HUMAN
|
||||||
| NC score | 0.042484 (rank : 32) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.036172 (rank : 33) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
K0319_HUMAN
|
||||||
| NC score | 0.034344 (rank : 34) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.032559 (rank : 35) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.031802 (rank : 36) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.027744 (rank : 37) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.025534 (rank : 38) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MMAC_HUMAN
|
||||||
| NC score | 0.024715 (rank : 39) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4U1, Q5T157, Q9BRQ7 | Gene names | MMACHC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methylmalonic aciduria and homocystinuria type C protein. | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.024606 (rank : 40) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.024160 (rank : 41) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.023201 (rank : 42) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CDC5L_MOUSE
|
||||||
| NC score | 0.022918 (rank : 43) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
CS018_HUMAN
|
||||||
| NC score | 0.021185 (rank : 44) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEA5 | Gene names | C19orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf18 precursor. | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.020196 (rank : 45) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
P66B_MOUSE
|
||||||
| NC score | 0.019927 (rank : 46) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHR5, Q8C9Q3 | Gene names | Gatad2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 beta (p66/p68) (GATA zinc finger domain- containing protein 2B). | |||||
|
CDC5L_HUMAN
|
||||||
| NC score | 0.019496 (rank : 47) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.018930 (rank : 48) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.018865 (rank : 49) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.018217 (rank : 50) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.017575 (rank : 51) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.017245 (rank : 52) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.017155 (rank : 53) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.017147 (rank : 54) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
MLTK_MOUSE
|
||||||
| NC score | 0.015198 (rank : 55) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.015034 (rank : 56) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | 0.014640 (rank : 57) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.012718 (rank : 58) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.011655 (rank : 59) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
DRBP1_HUMAN
|
||||||
| NC score | 0.011071 (rank : 60) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
I12R2_MOUSE
|
||||||
| NC score | 0.009694 (rank : 61) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97378 | Gene names | Il12rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
K6PF_HUMAN
|
||||||
| NC score | 0.009578 (rank : 62) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08237, Q16814, Q16815 | Gene names | PFKM | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructokinase, muscle type (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme A) (PFK-A) (Phosphofructokinase-M). | |||||
|
RFTN1_MOUSE
|
||||||
| NC score | 0.008840 (rank : 63) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A0D4, Q3U654, Q3U8P6, Q80US1 | Gene names | Rftn1, Kiaa0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.008389 (rank : 64) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.007634 (rank : 65) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.006441 (rank : 66) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.005824 (rank : 67) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.005597 (rank : 68) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.005096 (rank : 69) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.005004 (rank : 70) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
LATS2_MOUSE
|
||||||
| NC score | 0.004946 (rank : 71) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSJ6, Q8CDJ4, Q9JMI3 | Gene names | Lats2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.003712 (rank : 72) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.003686 (rank : 73) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | 0.003395 (rank : 74) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.003198 (rank : 75) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.001473 (rank : 76) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.001420 (rank : 77) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.000961 (rank : 78) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
ZN707_HUMAN
|
||||||
| NC score | -0.003271 (rank : 79) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96C28 | Gene names | ZNF707 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 707. | |||||