Please be patient as the page loads
|
S23IP_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S23IP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.913900 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 3) | NC score | 0.602223 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 4) | NC score | 0.609988 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.331219 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 6) | NC score | 0.322094 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
ANS4B_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 7) | NC score | 0.187578 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
PITM3_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 8) | NC score | 0.337311 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
USH1G_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 9) | NC score | 0.179356 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
USH1G_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 10) | NC score | 0.178794 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
ANS4B_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 11) | NC score | 0.181004 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
PITM3_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 12) | NC score | 0.329003 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 13) | NC score | 0.289572 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 14) | NC score | 0.288966 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
ANKS3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.099090 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.069356 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
ANKS3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.096516 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZW76, Q8TF25 | Gene names | ANKS3, KIAA1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.071327 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 19) | NC score | 0.068845 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.070715 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 21) | NC score | 0.091223 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.087583 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.019452 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.058795 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.105847 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.073941 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.095653 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.080663 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.017293 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.081026 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.082670 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.040184 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.028224 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.027608 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.059019 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.043820 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.048092 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.048589 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
DLX4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.017085 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.064602 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.031437 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.058939 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
EPHA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.010588 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.035329 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.056809 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.049725 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.022294 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EPHA5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.008676 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.051640 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.046015 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
LSP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.040299 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.081313 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SCMH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.032559 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
ALX3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.012523 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70137 | Gene names | Alx3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 3 (Proline-rich transcription factor ALX3). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.050453 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.074683 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.043020 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.014786 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.062807 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BOC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.008522 (rank : 166) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.030528 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EPHA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.009943 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60750, Q8CED9, Q9ESJ2 | Gene names | Epha1, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ESK). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.084611 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.027517 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
LMBL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.047065 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.051089 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.054374 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
SLIK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.007504 (rank : 169) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H156, Q5JXB1, Q8NBC7, Q96JH3 | Gene names | SLITRK2, CXorf2, KIAA1854, SLITL1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
BCL6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | -0.000105 (rank : 175) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.039733 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.039642 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.015241 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.049685 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.041771 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MISS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.046277 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.018097 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.022332 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.042950 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.025075 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.025308 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.072098 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.036036 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.036284 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.036405 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.036033 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.036270 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.036089 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.037341 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RT18B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.027759 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N84, Q3TLA9, Q9CRK0, Q9DCR8 | Gene names | Mrps18b | |||
|
Domain Architecture |
|
|||||
| Description | 28S ribosomal protein S18b, mitochondrial precursor (MRP-S18-b) (Mrps18b) (MRP-S18-2). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.011260 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
BTBD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.009654 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KE9, Q8IVQ7, Q9BR94 | Gene names | BTBD6, BDPL | |||
|
Domain Architecture |
|
|||||
| Description | BTB/POZ domain-containing protein 6 (Lens BTB domain protein). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.036822 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.019996 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
K1967_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.019068 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N163, Q6P0Q9, Q8N3G7, Q8N8M1, Q8TF34, Q9H9Q9, Q9HD12, Q9NT55 | Gene names | KIAA1967, DBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 (Deleted in breast cancer gene 1 protein) (DBC.1) (DBC-1) (p30 DBC). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.021277 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.014640 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.022399 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.010364 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
CLP46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.022586 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYB9, Q8R0H7 | Gene names | Clp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAP10-like 46 kDa protein precursor. | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.035815 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.035829 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.041700 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DMRTD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.017504 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGW9 | Gene names | Dmrtc2, Dmrt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2 (Doublesex- and mab-3-related transcription factor 7). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.026002 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
EPHA7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.006706 (rank : 171) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61772, Q61505, Q61773, Q61774 | Gene names | Epha7, Ebk, Ehk3, Mdk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 7 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-3) (EPH homology kinase 3) (Embryonic brain kinase) (EBK) (Developmental kinase 1) (MDK-1). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.023743 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.040361 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.062622 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.012064 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
CK024_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.036887 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CLP46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.021030 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBL1, Q53GJ4, Q8N2T1 | Gene names | CLP46, C3orf9, MDSRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAP10-like 46 kDa protein precursor (Myelodysplastic syndromes relative protein). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.018721 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
CTNA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.008127 (rank : 167) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI47, Q5VSR2, Q6P056 | Gene names | CTNNA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.047922 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.020834 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.014654 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MYO1E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.004568 (rank : 173) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.025809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.011032 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PITX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.007346 (rank : 170) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78337, O14677, O60425, Q9BTI5 | Gene names | PITX1, BFT, PTX1 | |||
|
Domain Architecture |
|
|||||
| Description | Pituitary homeobox 1 (Hindlimb expressed homeobox protein backfoot). | |||||
|
PITX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.007635 (rank : 168) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70314, O54742, P70427 | Gene names | Pitx1, Bft, Potx, Ptx1 | |||
|
Domain Architecture |
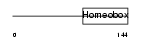
|
|||||
| Description | Pituitary homeobox 1 (Homeobox protein P-OTX) (Pituitary OTX-related factor) (Hindlimb expressed homeobox protein backfoot) (Ptx1). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.006657 (rank : 172) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.011483 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CCD76_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.012846 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH3 | Gene names | Ccdc76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 76. | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.014240 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.014245 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
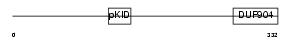
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.018806 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.015710 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
MLTK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.003479 (rank : 174) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.015428 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.016176 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.014126 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.029817 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.035339 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.067158 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050697 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051744 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CITE4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053818 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.064616 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.080031 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.056118 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.065815 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IASPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.057067 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.057891 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.071711 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.050247 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PIPNA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.112746 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |
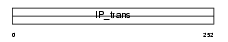
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.113214 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |
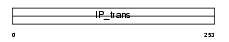
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.111446 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.111856 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.076527 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.076267 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.081229 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.069886 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.062487 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.065387 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.057945 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.059243 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058135 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.055506 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050415 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.050748 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.061184 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.059137 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.065525 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.055505 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055038 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.055455 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.075780 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.064551 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.064447 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.067230 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054441 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.052126 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.076902 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.913900 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
DDHD1_MOUSE
|
||||||
| NC score | 0.609988 (rank : 3) | θ value | 3.39556e-23 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.602223 (rank : 4) | θ value | 8.08199e-25 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
PITM3_MOUSE
|
||||||
| NC score | 0.337311 (rank : 5) | θ value | 2.21117e-06 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.331219 (rank : 6) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM3_HUMAN
|
||||||
| NC score | 0.329003 (rank : 7) | θ value | 1.43324e-05 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.322094 (rank : 8) | θ value | 2.36244e-08 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.289572 (rank : 9) | θ value | 7.1131e-05 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.288966 (rank : 10) | θ value | 0.000121331 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
ANS4B_MOUSE
|
||||||
| NC score | 0.187578 (rank : 11) | θ value | 5.81887e-07 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ANS4B_HUMAN
|
||||||
| NC score | 0.181004 (rank : 12) | θ value | 6.43352e-06 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
USH1G_MOUSE
|
||||||
| NC score | 0.179356 (rank : 13) | θ value | 2.21117e-06 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
USH1G_HUMAN
|
||||||
| NC score | 0.178794 (rank : 14) | θ value | 2.88788e-06 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
PIPNA_MOUSE
|
||||||
| NC score | 0.113214 (rank : 15) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_HUMAN
|
||||||
| NC score | 0.112746 (rank : 16) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_MOUSE
|
||||||
| NC score | 0.111856 (rank : 17) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_HUMAN
|
||||||
| NC score | 0.111446 (rank : 18) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.105847 (rank : 19) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
ANKS3_MOUSE
|
||||||
| NC score | 0.099090 (rank : 20) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
ANKS3_HUMAN
|
||||||
| NC score | 0.096516 (rank : 21) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZW76, Q8TF25 | Gene names | ANKS3, KIAA1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.095653 (rank : 22) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.091223 (rank : 23) | θ value | 0.00869519 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.087583 (rank : 24) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.084611 (rank : 25) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.082670 (rank : 26) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.081313 (rank : 27) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.081229 (rank : 28) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.081026 (rank : 29) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.080663 (rank : 30) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.080031 (rank : 31) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.076902 (rank : 32) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.076527 (rank : 33) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.076267 (rank : 34) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.075780 (rank : 35) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.074683 (rank : 36) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.073941 (rank : 37) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.072098 (rank : 38) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.071711 (rank : 39) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.071327 (rank : 40) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.070715 (rank : 41) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.069886 (rank : 42) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.069356 (rank : 43) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.068845 (rank : 44) | θ value | 0.00509761 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.067230 (rank : 45) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.067158 (rank : 46) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.065815 (rank : 47) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.065525 (rank : 48) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.065387 (rank : 49) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.064616 (rank : 50) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.064602 (rank : 51) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.064551 (rank : 52) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.064447 (rank : 53) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.062807 (rank : 54) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.062622 (rank : 55) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.062487 (rank : 56) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.061184 (rank : 57) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.059243 (rank : 58) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.059137 (rank : 59) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.059019 (rank : 60) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.058939 (rank : 61) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.058795 (rank : 62) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.058135 (rank : 63) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.057945 (rank : 64) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.057891 (rank : 65) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.057067 (rank : 66) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.056809 (rank : 67) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.056118 (rank : 68) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.055506 (rank : 69) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.055505 (rank : 70) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.055455 (rank : 71) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.055038 (rank : 72) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.054441 (rank : 73) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.054374 (rank : 74) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
CITE4_MOUSE
|
||||||
| NC score | 0.053818 (rank : 75) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.052126 (rank : 76) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.051744 (rank : 77) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.051640 (rank : 78) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.051089 (rank : 79) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.050748 (rank : 80) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.050697 (rank : 81) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.050453 (rank : 82) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.050415 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.050247 (rank : 84) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.049725 (rank : 85) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.049685 (rank : 86) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.048589 (rank : 87) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.048092 (rank : 88) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.047922 (rank : 89) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
LMBL3_HUMAN
|
||||||
| NC score | 0.047065 (rank : 90) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.046277 (rank : 91) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.046015 (rank : 92) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.043820 (rank : 93) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.043020 (rank : 94) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.042950 (rank : 95) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.041771 (rank : 96) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.041700 (rank : 97) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.040361 (rank : 98) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
LSP1_MOUSE
|
||||||
| NC score | 0.040299 (rank : 99) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.040184 (rank : 100) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.039733 (rank : 101) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.039642 (rank : 102) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.037341 (rank : 103) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.036887 (rank : 104) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.036822 (rank : 105) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.036405 (rank : 106) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.036284 (rank : 107) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.036270 (rank : 108) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.036089 (rank : 109) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.036036 (rank : 110) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.036033 (rank : 111) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.035829 (rank : 112) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.035815 (rank : 113) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.035339 (rank : 114) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.035329 (rank : 115) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SCMH1_HUMAN
|
||||||
| NC score | 0.032559 (rank : 116) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.031437 (rank : 117) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.030528 (rank : 118) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.029817 (rank : 119) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.028224 (rank : 120) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
RT18B_MOUSE
|
||||||
| NC score | 0.027759 (rank : 121) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N84, Q3TLA9, Q9CRK0, Q9DCR8 | Gene names | Mrps18b | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S18b, mitochondrial precursor (MRP-S18-b) (Mrps18b) (MRP-S18-2). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.027608 (rank : 122) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.027517 (rank : 123) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.026002 (rank : 124) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.025809 (rank : 125) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.025308 (rank : 126) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.025075 (rank : 127) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.023743 (rank : 128) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
CLP46_MOUSE
|
||||||
| NC score | 0.022586 (rank : 129) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYB9, Q8R0H7 | Gene names | Clp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAP10-like 46 kDa protein precursor. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.022399 (rank : 130) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.022332 (rank : 131) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.022294 (rank : 132) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.021277 (rank : 133) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CLP46_HUMAN
|
||||||
| NC score | 0.021030 (rank : 134) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBL1, Q53GJ4, Q8N2T1 | Gene names | CLP46, C3orf9, MDSRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAP10-like 46 kDa protein precursor (Myelodysplastic syndromes relative protein). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.020834 (rank : 135) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.019996 (rank : 136) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.019452 (rank : 137) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
K1967_HUMAN
|
||||||
| NC score | 0.019068 (rank : 138) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N163, Q6P0Q9, Q8N3G7, Q8N8M1, Q8TF34, Q9H9Q9, Q9HD12, Q9NT55 | Gene names | KIAA1967, DBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 (Deleted in breast cancer gene 1 protein) (DBC.1) (DBC-1) (p30 DBC). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.018806 (rank : 139) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.018721 (rank : 140) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.018097 (rank : 141) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
DMRTD_MOUSE
|
||||||
| NC score | 0.017504 (rank : 142) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGW9 | Gene names | Dmrtc2, Dmrt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2 (Doublesex- and mab-3-related transcription factor 7). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.017293 (rank : 143) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
DLX4_MOUSE
|
||||||
| NC score | 0.017085 (rank : 144) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.016176 (rank : 145) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.015710 (rank : 146) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.015428 (rank : 147) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.015241 (rank : 148) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.014786 (rank : 149) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.014654 (rank : 150) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.014640 (rank : 151) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.014245 (rank : 152) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.014240 (rank : 153) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.014126 (rank : 154) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CCD76_MOUSE
|
||||||
| NC score | 0.012846 (rank : 155) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH3 | Gene names | Ccdc76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 76. | |||||
|
ALX3_MOUSE
|
||||||
| NC score | 0.012523 (rank : 156) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70137 | Gene names | Alx3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 3 (Proline-rich transcription factor ALX3). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.012064 (rank : 157) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.011483 (rank : 158) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.011260 (rank : 159) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.011032 (rank : 160) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
EPHA1_HUMAN
|
||||||
| NC score | 0.010588 (rank : 161) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.010364 (rank : 162) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
EPHA1_MOUSE
|
||||||
| NC score | 0.009943 (rank : 163) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60750, Q8CED9, Q9ESJ2 | Gene names | Epha1, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ESK). | |||||
|
BTBD6_HUMAN
|
||||||
| NC score | 0.009654 (rank : 164) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KE9, Q8IVQ7, Q9BR94 | Gene names | BTBD6, BDPL | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 6 (Lens BTB domain protein). | |||||
|
EPHA5_HUMAN
|
||||||
| NC score | 0.008676 (rank : 165) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
BOC_HUMAN
|
||||||
| NC score | 0.008522 (rank : 166) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
CTNA3_HUMAN
|
||||||
| NC score | 0.008127 (rank : 167) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI47, Q5VSR2, Q6P056 | Gene names | CTNNA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
PITX1_MOUSE
|
||||||
| NC score | 0.007635 (rank : 168) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70314, O54742, P70427 | Gene names | Pitx1, Bft, Potx, Ptx1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 1 (Homeobox protein P-OTX) (Pituitary OTX-related factor) (Hindlimb expressed homeobox protein backfoot) (Ptx1). | |||||
|
SLIK2_HUMAN
|
||||||
| NC score | 0.007504 (rank : 169) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H156, Q5JXB1, Q8NBC7, Q96JH3 | Gene names | SLITRK2, CXorf2, KIAA1854, SLITL1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
PITX1_HUMAN
|
||||||
| NC score | 0.007346 (rank : 170) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78337, O14677, O60425, Q9BTI5 | Gene names | PITX1, BFT, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 1 (Hindlimb expressed homeobox protein backfoot). | |||||
|
EPHA7_MOUSE
|
||||||
| NC score | 0.006706 (rank : 171) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61772, Q61505, Q61773, Q61774 | Gene names | Epha7, Ebk, Ehk3, Mdk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 7 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-3) (EPH homology kinase 3) (Embryonic brain kinase) (EBK) (Developmental kinase 1) (MDK-1). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.006657 (rank : 172) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
MYO1E_HUMAN
|
||||||
| NC score | 0.004568 (rank : 173) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
MLTK_MOUSE
|
||||||
| NC score | 0.003479 (rank : 174) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
BCL6_MOUSE
|
||||||
| NC score | -0.000105 (rank : 175) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||