Please be patient as the page loads
|
PITM1_HUMAN
|
||||||
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PITM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989541 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.977353 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.972606 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.886809 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.887771 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PIPNB_MOUSE
|
||||||
| θ value | 4.53632e-60 (rank : 7) | NC score | 0.771277 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |
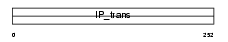
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNA_MOUSE
|
||||||
| θ value | 1.31987e-59 (rank : 8) | NC score | 0.775170 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |
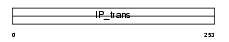
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 9) | NC score | 0.774060 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |
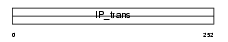
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 10) | NC score | 0.770502 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |
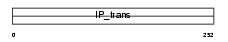
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
DDHD1_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 11) | NC score | 0.372731 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 12) | NC score | 0.322094 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 13) | NC score | 0.359202 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 14) | NC score | 0.317692 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.020302 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.023125 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.035167 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.028062 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
HXC4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.015037 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09017 | Gene names | HOXC4, HOX3E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C4 (Hox-3E) (CP19). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.042619 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.032043 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.028096 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.041183 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
HXC4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.013838 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08624 | Gene names | Hoxc4, Hox-3.5, Hoxc-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C4 (Hox-3.5). | |||||
|
LRC46_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.023708 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FV0 | Gene names | LRRC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 46. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.028340 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.036867 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.024820 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.017268 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.004028 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.017320 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.019266 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.036552 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.028588 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.025452 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HXA4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.011577 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.026383 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KLH14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.006646 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
PAX9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.022406 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.010069 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.035971 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TTC15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.023398 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
CD5R2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.019580 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.008711 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.027029 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.020755 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.025019 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.019550 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.023859 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
VNN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.017816 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95497, Q4JFW6, Q4VAS7, Q4VAS8, Q4VAS9, Q9UF16, Q9UJF4 | Gene names | VNN1 | |||
|
Domain Architecture |
|
|||||
| Description | Pantetheinase precursor (EC 3.5.1.92) (Pantetheine hydrolase) (Vascular non-inflammatory molecule 1) (Vanin-1) (Tiff66). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.011977 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.025180 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.020108 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.023700 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.014869 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.034266 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
ZN282_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | -0.000687 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.021261 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
ATS19_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.004376 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE59 | Gene names | ADAMTS19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.014322 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.015968 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.008896 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.018705 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.004376 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
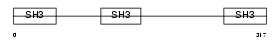
CD2AP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.015490 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |
|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
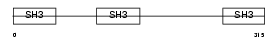
CD2AP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.015058 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.016377 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.015193 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.007393 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.022793 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.023013 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
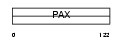
PAX9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.018450 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.009322 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
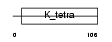
ZBT17_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | -0.001660 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.014518 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.011516 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.013370 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.024265 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.018528 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.014443 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.009483 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.013455 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PO2F3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.006857 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31362 | Gene names | Pou2f3, Epoc1, Oct11, Otf-11, Otf11 | |||
|
Domain Architecture |
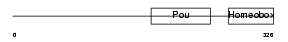
|
|||||
| Description | POU domain, class 2, transcription factor 3 (Octamer-binding transcription factor 11) (Oct-11) (Epoc-1). | |||||
|
SETB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.011094 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.012980 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.005688 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CRX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.005374 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |
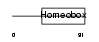
|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
FA35A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.017867 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
HXA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.009891 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00056, O43366 | Gene names | HOXA4, HOX1D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1D) (Hox-1.4). | |||||
|
KLF14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | -0.001237 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.017028 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.019773 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.013330 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
S2A4R_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.018120 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR83, Q6F6I6, Q6F6I7, Q6GTK5, Q8TAH5, Q8WVW7, Q96QD3, Q9BV85 | Gene names | SLC2A4RG, HDBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLC2A4 regulator (GLUT4 enhancer factor) (GEF) (Huntington disease gene regulatory region-binding protein 1) (HDBP-1). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.008400 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.016851 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.006525 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
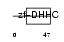
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
DEND_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.011297 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.001601 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
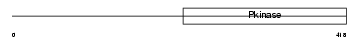
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.015242 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.004100 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.009955 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.007075 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.005440 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.018958 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.008814 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
SPSY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.020759 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52788, O00544, Q9UQS1 | Gene names | SMS | |||
|
Domain Architecture |

|
|||||
| Description | Spermine synthase (EC 2.5.1.22) (Spermidine aminopropyltransferase) (SPMSY). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.017508 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.026272 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
VEGFB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.009656 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
ZFPL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.011578 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB43 | Gene names | Zfpl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein-like 1. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.010607 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.011655 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.008929 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.013413 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CPT1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.003596 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924X2, O35287, Q9QYP4 | Gene names | Cpt1b | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-palmitoyltransferase I, muscle isoform (EC 2.3.1.21) (CPT I) (CPTI-M) (Carnitine palmitoyltransferase 1B). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.016005 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.007814 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07820, Q9HD91, Q9NRQ3, Q9NRQ4, Q9UHR7, Q9UHR8, Q9UHR9, Q9UNJ1 | Gene names | MCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 (Bcl-2- related protein EAT/mcl1) (mcl1/EAT). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.015990 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.014318 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.011464 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.008905 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.008506 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SHC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.003649 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.007031 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SPSY_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.018363 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97355, Q3TL65 | Gene names | Sms | |||
|
Domain Architecture |

|
|||||
| Description | Spermine synthase (EC 2.5.1.22) (Spermidine aminopropyltransferase) (SPMSY). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.016308 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.009827 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | -0.000371 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.989541 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.977353 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.972606 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM3_MOUSE
|
||||||
| NC score | 0.887771 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_HUMAN
|
||||||
| NC score | 0.886809 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PIPNA_MOUSE
|
||||||
| NC score | 0.775170 (rank : 7) | θ value | 1.31987e-59 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_HUMAN
|
||||||
| NC score | 0.774060 (rank : 8) | θ value | 8.55514e-59 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_MOUSE
|
||||||
| NC score | 0.771277 (rank : 9) | θ value | 4.53632e-60 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_HUMAN
|
||||||
| NC score | 0.770502 (rank : 10) | θ value | 2.48917e-58 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
DDHD1_MOUSE
|
||||||
| NC score | 0.372731 (rank : 11) | θ value | 1.38499e-08 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.359202 (rank : 12) | θ value | 4.0297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.322094 (rank : 13) | θ value | 2.36244e-08 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.317692 (rank : 14) | θ value | 4.0297e-08 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.042619 (rank : 15) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.041183 (rank : 16) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.036867 (rank : 17) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.036552 (rank : 18) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.035971 (rank : 19) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.035167 (rank : 20) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.034266 (rank : 21) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.032043 (rank : 22) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.028588 (rank : 23) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.028340 (rank : 24) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.028096 (rank : 25) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.028062 (rank : 26) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.027029 (rank : 27) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.026383 (rank : 28) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.026272 (rank : 29) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.025452 (rank : 30) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.025180 (rank : 31) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.025019 (rank : 32) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.024820 (rank : 33) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.024265 (rank : 34) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.023859 (rank : 35) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
LRC46_HUMAN
|
||||||
| NC score | 0.023708 (rank : 36) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FV0 | Gene names | LRRC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 46. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.023700 (rank : 37) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TTC15_HUMAN
|
||||||
| NC score | 0.023398 (rank : 38) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.023125 (rank : 39) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.023013 (rank : 40) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.022793 (rank : 41) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PAX9_MOUSE
|
||||||
| NC score | 0.022406 (rank : 42) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.021261 (rank : 43) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
SPSY_HUMAN
|
||||||
| NC score | 0.020759 (rank : 44) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52788, O00544, Q9UQS1 | Gene names | SMS | |||
|
Domain Architecture |

|
|||||
| Description | Spermine synthase (EC 2.5.1.22) (Spermidine aminopropyltransferase) (SPMSY). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.020755 (rank : 45) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.020302 (rank : 46) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.020108 (rank : 47) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.019773 (rank : 48) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CD5R2_MOUSE
|
||||||
| NC score | 0.019580 (rank : 49) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.019550 (rank : 50) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.019266 (rank : 51) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.018958 (rank : 52) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.018705 (rank : 53) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.018528 (rank : 54) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PAX9_HUMAN
|
||||||
| NC score | 0.018450 (rank : 55) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
SPSY_MOUSE
|
||||||
| NC score | 0.018363 (rank : 56) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97355, Q3TL65 | Gene names | Sms | |||
|
Domain Architecture |

|
|||||
| Description | Spermine synthase (EC 2.5.1.22) (Spermidine aminopropyltransferase) (SPMSY). | |||||
|
S2A4R_HUMAN
|
||||||
| NC score | 0.018120 (rank : 57) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR83, Q6F6I6, Q6F6I7, Q6GTK5, Q8TAH5, Q8WVW7, Q96QD3, Q9BV85 | Gene names | SLC2A4RG, HDBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLC2A4 regulator (GLUT4 enhancer factor) (GEF) (Huntington disease gene regulatory region-binding protein 1) (HDBP-1). | |||||
|
FA35A_MOUSE
|
||||||
| NC score | 0.017867 (rank : 58) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
VNN1_HUMAN
|
||||||
| NC score | 0.017816 (rank : 59) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95497, Q4JFW6, Q4VAS7, Q4VAS8, Q4VAS9, Q9UF16, Q9UJF4 | Gene names | VNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Pantetheinase precursor (EC 3.5.1.92) (Pantetheine hydrolase) (Vascular non-inflammatory molecule 1) (Vanin-1) (Tiff66). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.017508 (rank : 60) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.017320 (rank : 61) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.017268 (rank : 62) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.017028 (rank : 63) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.016851 (rank : 64) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.016377 (rank : 65) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.016308 (rank : 66) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.016005 (rank : 67) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.015990 (rank : 68) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.015968 (rank : 69) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
CD2AP_HUMAN
|
||||||
| NC score | 0.015490 (rank : 70) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.015242 (rank : 71) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.015193 (rank : 72) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.015058 (rank : 73) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
HXC4_HUMAN
|
||||||
| NC score | 0.015037 (rank : 74) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09017 | Gene names | HOXC4, HOX3E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C4 (Hox-3E) (CP19). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.014869 (rank : 75) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.014518 (rank : 76) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.014443 (rank : 77) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.014322 (rank : 78) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.014318 (rank : 79) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
HXC4_MOUSE
|
||||||
| NC score | 0.013838 (rank : 80) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08624 | Gene names | Hoxc4, Hox-3.5, Hoxc-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C4 (Hox-3.5). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.013455 (rank : 81) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.013413 (rank : 82) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.013370 (rank : 83) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.013330 (rank : 84) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.012980 (rank : 85) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.011977 (rank : 86) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.011655 (rank : 87) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ZFPL1_MOUSE
|
||||||
| NC score | 0.011578 (rank : 88) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB43 | Gene names | Zfpl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein-like 1. | |||||
|
HXA4_MOUSE
|
||||||
| NC score | 0.011577 (rank : 89) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.011516 (rank : 90) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.011464 (rank : 91) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.011297 (rank : 92) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
SETB1_HUMAN
|
||||||
| NC score | 0.011094 (rank : 93) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.010607 (rank : 94) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.010069 (rank : 95) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.009955 (rank : 96) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
HXA4_HUMAN
|
||||||
| NC score | 0.009891 (rank : 97) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00056, O43366 | Gene names | HOXA4, HOX1D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1D) (Hox-1.4). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.009827 (rank : 98) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
VEGFB_HUMAN
|
||||||
| NC score | 0.009656 (rank : 99) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.009483 (rank : 100) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.009322 (rank : 101) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.008929 (rank : 102) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.008905 (rank : 103) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.008896 (rank : 104) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.008814 (rank : 105) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.008711 (rank : 106) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.008506 (rank : 107) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.008400 (rank : 108) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
MCL1_HUMAN
|
||||||
| NC score | 0.007814 (rank : 109) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07820, Q9HD91, Q9NRQ3, Q9NRQ4, Q9UHR7, Q9UHR8, Q9UHR9, Q9UNJ1 | Gene names | MCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 (Bcl-2- related protein EAT/mcl1) (mcl1/EAT). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.007393 (rank : 110) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.007075 (rank : 111) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.007031 (rank : 112) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
PO2F3_MOUSE
|
||||||
| NC score | 0.006857 (rank : 113) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31362 | Gene names | Pou2f3, Epoc1, Oct11, Otf-11, Otf11 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 3 (Octamer-binding transcription factor 11) (Oct-11) (Epoc-1). | |||||
|
KLH14_HUMAN
|
||||||
| NC score | 0.006646 (rank : 114) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.006525 (rank : 115) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.005688 (rank : 116) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.005440 (rank : 117) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
CRX_MOUSE
|
||||||
| NC score | 0.005374 (rank : 118) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |

|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
ATS19_HUMAN
|
||||||
| NC score | 0.004376 (rank : 119) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE59 | Gene names | ADAMTS19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.004376 (rank : 120) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.004100 (rank : 121) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.004028 (rank : 122) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
SHC1_MOUSE
|
||||||
| NC score | 0.003649 (rank : 123) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
CPT1B_MOUSE
|
||||||
| NC score | 0.003596 (rank : 124) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924X2, O35287, Q9QYP4 | Gene names | Cpt1b | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-palmitoyltransferase I, muscle isoform (EC 2.3.1.21) (CPT I) (CPTI-M) (Carnitine palmitoyltransferase 1B). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.001601 (rank : 125) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
ZN219_HUMAN
|
||||||
| NC score | -0.000371 (rank : 126) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
ZN282_HUMAN
|
||||||
| NC score | -0.000687 (rank : 127) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
KLF14_HUMAN
|
||||||
| NC score | -0.001237 (rank : 128) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | -0.001660 (rank : 129) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||