Please be patient as the page loads
|
RCOR3_MOUSE
|
||||||
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RCOR3_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 1.97579e-164 (rank : 2) | NC score | 0.973363 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 3.20603e-106 (rank : 3) | NC score | 0.957749 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 6.0463e-105 (rank : 4) | NC score | 0.954482 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 7.15887e-98 (rank : 5) | NC score | 0.956988 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 9.34977e-98 (rank : 6) | NC score | 0.953945 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 7) | NC score | 0.511326 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
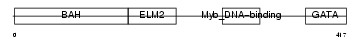
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 8) | NC score | 0.502558 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 9) | NC score | 0.529368 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 10) | NC score | 0.398857 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 11) | NC score | 0.397585 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 12) | NC score | 0.499093 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 13) | NC score | 0.500286 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
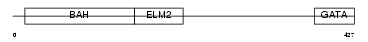
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 14) | NC score | 0.470958 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
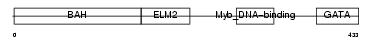
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 15) | NC score | 0.466434 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
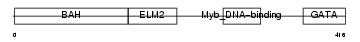
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 16) | NC score | 0.399890 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.454966 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.451061 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.383545 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 20) | NC score | 0.386781 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 21) | NC score | 0.351803 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 22) | NC score | 0.381758 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.348903 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 24) | NC score | 0.109840 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.082475 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 26) | NC score | 0.073140 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.063587 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.215813 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.059483 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.216448 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 31) | NC score | 0.081320 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.084736 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.045631 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
RED_MOUSE
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.083938 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.055657 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.029857 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
RED_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.079851 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
CJ108_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.090038 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.041274 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.024262 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.055258 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.042373 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.050141 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.043376 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PLK3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | -0.000318 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.011537 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
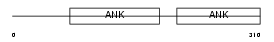
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.024184 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.034542 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.042309 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.057749 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.030980 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.017884 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.035404 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.033969 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.039673 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.047554 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.025062 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.047310 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.035095 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CNOT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.041284 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |
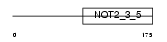
|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CNOT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.041565 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.035060 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.065316 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.014869 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.060399 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.053256 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.054638 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.015309 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.029719 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.025999 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
LPIN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.016861 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
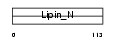
|
|||||
| Description | Lipin-3. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.002426 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.041553 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.020479 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
CJ086_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.037034 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.029462 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.066072 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.040245 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.056284 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.032282 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.053429 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZN342_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | -0.000581 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.005096 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.010659 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.002010 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
P73_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.021748 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.014482 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.015223 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.053405 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.002982 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
NOC2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.019871 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.040109 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
QSCN6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.014443 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.007510 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.029242 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.006010 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.010197 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.010279 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.016959 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.004063 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.020011 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015170 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.010927 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.052435 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008175 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.009865 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.012866 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
P66A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.016407 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004998 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014044 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.089514 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.058862 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051816 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055459 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054102 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.973363 (rank : 2) | θ value | 1.97579e-164 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.957749 (rank : 3) | θ value | 3.20603e-106 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.956988 (rank : 4) | θ value | 7.15887e-98 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.954482 (rank : 5) | θ value | 6.0463e-105 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.953945 (rank : 6) | θ value | 9.34977e-98 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.529368 (rank : 7) | θ value | 3.29651e-10 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.511326 (rank : 8) | θ value | 1.133e-10 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.502558 (rank : 9) | θ value | 2.52405e-10 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.500286 (rank : 10) | θ value | 9.59137e-10 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.499093 (rank : 11) | θ value | 5.62301e-10 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.470958 (rank : 12) | θ value | 4.0297e-08 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.466434 (rank : 13) | θ value | 5.26297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.454966 (rank : 14) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.451061 (rank : 15) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.399890 (rank : 16) | θ value | 1.69304e-06 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.398857 (rank : 17) | θ value | 4.30538e-10 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.397585 (rank : 18) | θ value | 4.30538e-10 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.386781 (rank : 19) | θ value | 3.19293e-05 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.383545 (rank : 20) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.381758 (rank : 21) | θ value | 4.1701e-05 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.351803 (rank : 22) | θ value | 3.19293e-05 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.348903 (rank : 23) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.216448 (rank : 24) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.215813 (rank : 25) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.109840 (rank : 26) | θ value | 0.000158464 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.090038 (rank : 27) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.089514 (rank : 28) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.084736 (rank : 29) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
RED_MOUSE
|
||||||
| NC score | 0.083938 (rank : 30) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.082475 (rank : 31) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.081320 (rank : 32) | θ value | 0.0330416 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
RED_HUMAN
|
||||||
| NC score | 0.079851 (rank : 33) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.073140 (rank : 34) | θ value | 0.00298849 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.066072 (rank : 35) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.065316 (rank : 36) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.063587 (rank : 37) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.060399 (rank : 38) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.059483 (rank : 39) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058862 (rank : 40) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.057749 (rank : 41) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.056284 (rank : 42) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.055657 (rank : 43) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.055459 (rank : 44) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.055258 (rank : 45) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.054638 (rank : 46) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.054102 (rank : 47) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.053429 (rank : 48) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.053405 (rank : 49) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.053256 (rank : 50) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.052435 (rank : 51) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.051816 (rank : 52) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.050141 (rank : 53) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.047554 (rank : 54) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.047310 (rank : 55) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.045631 (rank : 56) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.043376 (rank : 57) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.042373 (rank : 58) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.042309 (rank : 59) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
CNOT2_MOUSE
|
||||||
| NC score | 0.041565 (rank : 60) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.041553 (rank : 61) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
CNOT2_HUMAN
|
||||||
| NC score | 0.041284 (rank : 62) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.041274 (rank : 63) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.040245 (rank : 64) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.040109 (rank : 65) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.039673 (rank : 66) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
CJ086_HUMAN
|
||||||
| NC score | 0.037034 (rank : 67) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.035404 (rank : 68) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.035095 (rank : 69) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.035060 (rank : 70) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.034542 (rank : 71) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.033969 (rank : 72) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.032282 (rank : 73) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.030980 (rank : 74) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.029857 (rank : 75) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.029719 (rank : 76) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.029462 (rank : 77) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.029242 (rank : 78) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.025999 (rank : 79) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.025062 (rank : 80) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.024262 (rank : 81) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.024184 (rank : 82) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.021748 (rank : 83) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.020479 (rank : 84) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.020011 (rank : 85) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.019871 (rank : 86) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.017884 (rank : 87) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.016959 (rank : 88) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.016861 (rank : 89) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.016407 (rank : 90) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.015309 (rank : 91) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.015223 (rank : 92) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.015170 (rank : 93) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.014869 (rank : 94) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.014482 (rank : 95) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
QSCN6_HUMAN
|
||||||
| NC score | 0.014443 (rank : 96) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.014044 (rank : 97) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.012866 (rank : 98) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.011537 (rank : 99) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.010927 (rank : 100) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.010659 (rank : 101) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.010279 (rank : 102) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.010197 (rank : 103) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.009865 (rank : 104) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.008175 (rank : 105) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.007510 (rank : 106) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.006010 (rank : 107) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.005096 (rank : 108) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.004998 (rank : 109) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.004063 (rank : 110) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.002982 (rank : 111) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.002426 (rank : 112) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.002010 (rank : 113) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PLK3_HUMAN
|
||||||
| NC score | -0.000318 (rank : 114) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
ZN342_HUMAN
|
||||||
| NC score | -0.000581 (rank : 115) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||