Please be patient as the page loads
|
MTA2_HUMAN
|
||||||
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990179 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
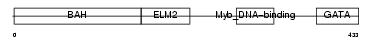
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989771 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
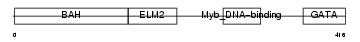
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999074 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991000 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
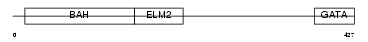
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.991101 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
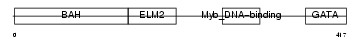
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 7) | NC score | 0.593248 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.32485e-38 (rank : 8) | NC score | 0.587595 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 9) | NC score | 0.530602 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 10) | NC score | 0.525135 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 11) | NC score | 0.522788 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 12) | NC score | 0.478327 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 13) | NC score | 0.477229 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 14) | NC score | 0.470551 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 15) | NC score | 0.502558 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 16) | NC score | 0.512251 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 17) | NC score | 0.281565 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 18) | NC score | 0.281144 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.340873 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.019125 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.136591 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.038849 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.038937 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.057415 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.017208 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.035135 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
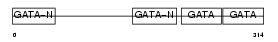
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.016493 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.039565 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
AT5G1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.011970 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48202 | Gene names | Atp5g1 | |||
|
Domain Architecture |
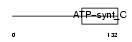
|
|||||
| Description | ATP synthase lipid-binding protein, mitochondrial precursor (EC 3.6.3.14) (ATP synthase proteolipid P1) (ATPase protein 9) (ATPase subunit C). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.020005 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.007984 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.004908 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.005217 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.005777 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
CAH5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.002970 (rank : 43) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35218 | Gene names | CA5A, CA5 | |||
|
Domain Architecture |
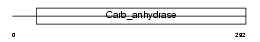
|
|||||
| Description | Carbonic anhydrase 5A, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VA) (Carbonate dehydratase VA) (CA-VA). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.029414 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
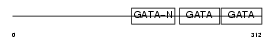
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.031828 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.005143 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.003713 (rank : 42) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.135237 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.089057 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.090574 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.093441 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.999074 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.991101 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.991000 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.990179 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.989771 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.593248 (rank : 7) | θ value | 2.86122e-38 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.587595 (rank : 8) | θ value | 8.32485e-38 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.530602 (rank : 9) | θ value | 4.16044e-13 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.525135 (rank : 10) | θ value | 7.09661e-13 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.522788 (rank : 11) | θ value | 1.133e-10 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.512251 (rank : 12) | θ value | 5.62301e-10 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.502558 (rank : 13) | θ value | 2.52405e-10 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.478327 (rank : 14) | θ value | 2.52405e-10 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.477229 (rank : 15) | θ value | 2.52405e-10 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.470551 (rank : 16) | θ value | 2.52405e-10 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.340873 (rank : 17) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.281565 (rank : 18) | θ value | 4.1701e-05 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.281144 (rank : 19) | θ value | 0.00035302 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.136591 (rank : 20) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.135237 (rank : 21) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.093441 (rank : 22) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.090574 (rank : 23) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.089057 (rank : 24) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.057415 (rank : 25) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.039565 (rank : 26) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.038937 (rank : 27) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.038849 (rank : 28) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.035135 (rank : 29) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.031828 (rank : 30) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.029414 (rank : 31) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.020005 (rank : 32) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.019125 (rank : 33) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.017208 (rank : 34) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.016493 (rank : 35) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
AT5G1_MOUSE
|
||||||
| NC score | 0.011970 (rank : 36) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48202 | Gene names | Atp5g1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase lipid-binding protein, mitochondrial precursor (EC 3.6.3.14) (ATP synthase proteolipid P1) (ATPase protein 9) (ATPase subunit C). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.007984 (rank : 37) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.005777 (rank : 38) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.005217 (rank : 39) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.005143 (rank : 40) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.004908 (rank : 41) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.003713 (rank : 42) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
CAH5A_HUMAN
|
||||||
| NC score | 0.002970 (rank : 43) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35218 | Gene names | CA5A, CA5 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 5A, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VA) (Carbonate dehydratase VA) (CA-VA). | |||||