Please be patient as the page loads
|
GATA4_MOUSE
|
||||||
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
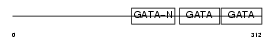
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GATA4_MOUSE
|
||||||
| θ value | 1.36511e-173 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 1.24331e-150 (rank : 2) | NC score | 0.994220 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
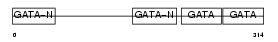
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 2.24094e-75 (rank : 3) | NC score | 0.969319 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| θ value | 8.51556e-75 (rank : 4) | NC score | 0.970254 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 5) | NC score | 0.971123 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 2.03155e-68 (rank : 6) | NC score | 0.970866 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA2_HUMAN
|
||||||
| θ value | 8.85319e-56 (rank : 7) | NC score | 0.939810 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA2_MOUSE
|
||||||
| θ value | 3.36421e-55 (rank : 8) | NC score | 0.936946 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA3_MOUSE
|
||||||
| θ value | 3.36421e-55 (rank : 9) | NC score | 0.946092 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| θ value | 1.66964e-54 (rank : 10) | NC score | 0.944687 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA1_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 11) | NC score | 0.957379 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA1_HUMAN
|
||||||
| θ value | 1.91031e-50 (rank : 12) | NC score | 0.954998 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 13) | NC score | 0.487610 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 14) | NC score | 0.478044 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.029304 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.027028 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.023793 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |
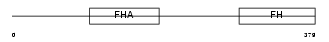
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.016145 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.016800 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.013792 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.058036 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.014547 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ZN683_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.005337 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
ATRIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.021500 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.015222 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.051231 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.012671 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
P66A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.042052 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
ACM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | -0.000048 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 930 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08172, Q9P1X9 | Gene names | CHRM2 | |||
|
Domain Architecture |
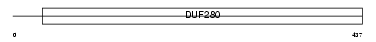
|
|||||
| Description | Muscarinic acetylcholine receptor M2. | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.005308 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.023094 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.007607 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.000556 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.015941 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.012818 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.014790 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
FOXH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.012086 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.007594 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.011225 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CDX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.003458 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14627, Q5JS20 | Gene names | CDX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.002335 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.005386 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.013020 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO002_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.009530 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
DLX4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.001389 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.014645 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
IGSF8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.006535 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969P0, Q8NG09, Q96DP4, Q9BTG9 | Gene names | IGSF8, CD81P3, EWI2, KCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 8 precursor (CD81 partner 3) (Glu- Trp-Ile EWI motif-containing protein 2) (EWI-2) (Keratinocytes- associated transmembrane protein 4) (KCT-4) (LIR-D1) (CD316 antigen). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.029414 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.029375 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.007164 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.009629 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.012820 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.36511e-173 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.994220 (rank : 2) | θ value | 1.24331e-150 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.971123 (rank : 3) | θ value | 1.83746e-69 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.970866 (rank : 4) | θ value | 2.03155e-68 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.970254 (rank : 5) | θ value | 8.51556e-75 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.969319 (rank : 6) | θ value | 2.24094e-75 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA1_MOUSE
|
||||||
| NC score | 0.957379 (rank : 7) | θ value | 1.72781e-51 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA1_HUMAN
|
||||||
| NC score | 0.954998 (rank : 8) | θ value | 1.91031e-50 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA3_MOUSE
|
||||||
| NC score | 0.946092 (rank : 9) | θ value | 3.36421e-55 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| NC score | 0.944687 (rank : 10) | θ value | 1.66964e-54 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA2_HUMAN
|
||||||
| NC score | 0.939810 (rank : 11) | θ value | 8.85319e-56 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA2_MOUSE
|
||||||
| NC score | 0.936946 (rank : 12) | θ value | 3.36421e-55 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.487610 (rank : 13) | θ value | 6.41864e-14 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.478044 (rank : 14) | θ value | 2.43908e-13 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.058036 (rank : 15) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.051231 (rank : 16) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.042052 (rank : 17) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.029414 (rank : 18) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.029375 (rank : 19) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.029304 (rank : 20) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.027028 (rank : 21) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.023793 (rank : 22) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.023094 (rank : 23) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
ATRIP_MOUSE
|
||||||
| NC score | 0.021500 (rank : 24) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.016800 (rank : 25) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.016145 (rank : 26) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.015941 (rank : 27) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.015222 (rank : 28) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.014790 (rank : 29) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.014645 (rank : 30) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.014547 (rank : 31) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.013792 (rank : 32) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.013020 (rank : 33) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.012820 (rank : 34) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.012818 (rank : 35) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.012671 (rank : 36) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
FOXH1_MOUSE
|
||||||
| NC score | 0.012086 (rank : 37) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.011225 (rank : 38) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.009629 (rank : 39) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.009530 (rank : 40) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.007607 (rank : 41) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.007594 (rank : 42) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.007164 (rank : 43) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
IGSF8_HUMAN
|
||||||
| NC score | 0.006535 (rank : 44) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969P0, Q8NG09, Q96DP4, Q9BTG9 | Gene names | IGSF8, CD81P3, EWI2, KCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 8 precursor (CD81 partner 3) (Glu- Trp-Ile EWI motif-containing protein 2) (EWI-2) (Keratinocytes- associated transmembrane protein 4) (KCT-4) (LIR-D1) (CD316 antigen). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.005386 (rank : 45) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
ZN683_HUMAN
|
||||||
| NC score | 0.005337 (rank : 46) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.005308 (rank : 47) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CDX4_HUMAN
|
||||||
| NC score | 0.003458 (rank : 48) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14627, Q5JS20 | Gene names | CDX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.002335 (rank : 49) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
DLX4_MOUSE
|
||||||
| NC score | 0.001389 (rank : 50) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.000556 (rank : 51) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ACM2_HUMAN
|
||||||
| NC score | -0.000048 (rank : 52) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 930 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08172, Q9P1X9 | Gene names | CHRM2 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M2. | |||||