Please be patient as the page loads
|
CAN7_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAN7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994397 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN5_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 3) | NC score | 0.763843 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 4) | NC score | 0.760184 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 5) | NC score | 0.750212 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 6) | NC score | 0.750222 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 7) | NC score | 0.666878 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 8) | NC score | 0.673186 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 9) | NC score | 0.671496 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 10) | NC score | 0.739659 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN11_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 11) | NC score | 0.676980 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 12) | NC score | 0.671336 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN1_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 13) | NC score | 0.662650 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 14) | NC score | 0.669208 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN10_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 15) | NC score | 0.733858 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN12_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 16) | NC score | 0.695444 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 17) | NC score | 0.644492 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN12_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 18) | NC score | 0.684473 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 19) | NC score | 0.644967 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CF103_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 20) | NC score | 0.408685 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.093787 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 22) | NC score | 0.095988 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.092388 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 24) | NC score | 0.089855 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
KS6C1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.027220 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.038343 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.011597 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.023450 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
KS6C1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.024532 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.023589 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.051231 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
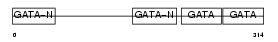
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.051231 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
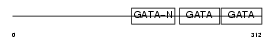
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.018191 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.050271 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
ULK3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.009282 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3U3Q1, Q8K1X6, Q9DBR8 | Gene names | Ulk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK3 (EC 2.7.11.1) (Unc-51-like kinase 3). | |||||
|
FBLN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.006577 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.047817 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.048044 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.008391 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.016495 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.009371 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TPSNR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.020113 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
VPS18_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.029131 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.044712 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
K0146_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.035217 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.012609 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.011962 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.010698 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.012635 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RPKL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.016674 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6S9, Q69YT9, Q6ZMQ6, Q96NR9, Q9BSU9 | Gene names | RPS6KL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
VPS18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.026957 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
FLNB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.009031 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.010495 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.010585 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013336 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RT09_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.030006 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82933 | Gene names | MRPS9, RPMS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S9, mitochondrial precursor (S9mt) (MRP-S9). | |||||
|
SNX15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.051628 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.010735 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ARI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.011812 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.011807 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
FBX9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.013469 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK97, O75986, Q6PKH7, Q9NT57, Q9Y593 | Gene names | FBXO9, FBX9, VCIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 9 (NY-REN-57 antigen) (Cross-immune reaction antigen 1). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.014163 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.011734 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.018281 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.016667 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.011506 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CERU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.008422 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
FBLN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.004494 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |
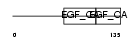
|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.005793 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.008031 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007863 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.054818 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CPNS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.331789 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.334228 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.329377 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.326712 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.251495 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
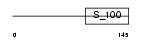
|
|||||
| Description | Grancalcin. | |||||
|
GRAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.257145 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.193146 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
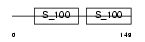
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.192231 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
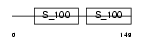
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.230543 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.237151 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
SORCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.252904 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
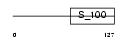
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.255023 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.994397 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN5_MOUSE
|
||||||
| NC score | 0.763843 (rank : 3) | θ value | 3.50572e-28 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| NC score | 0.760184 (rank : 4) | θ value | 3.28125e-26 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_MOUSE
|
||||||
| NC score | 0.750222 (rank : 5) | θ value | 1.86321e-21 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.750212 (rank : 6) | θ value | 1.42661e-21 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.739659 (rank : 7) | θ value | 5.07402e-19 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_MOUSE
|
||||||
| NC score | 0.733858 (rank : 8) | θ value | 1.63225e-17 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN12_HUMAN
|
||||||
| NC score | 0.695444 (rank : 9) | θ value | 1.63225e-17 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| NC score | 0.684473 (rank : 10) | θ value | 2.20605e-14 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.676980 (rank : 11) | θ value | 1.13037e-18 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.673186 (rank : 12) | θ value | 3.88503e-19 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.671496 (rank : 13) | θ value | 3.88503e-19 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.671336 (rank : 14) | θ value | 1.92812e-18 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.669208 (rank : 15) | θ value | 9.56915e-18 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.666878 (rank : 16) | θ value | 1.02238e-19 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_MOUSE
|
||||||
| NC score | 0.662650 (rank : 17) | θ value | 9.56915e-18 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.644967 (rank : 18) | θ value | 2.88119e-14 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.644492 (rank : 19) | θ value | 9.90251e-15 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.408685 (rank : 20) | θ value | 2.0648e-12 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
CPNS1_MOUSE
|
||||||
| NC score | 0.334228 (rank : 21) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| NC score | 0.331789 (rank : 22) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.329377 (rank : 23) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.326712 (rank : 24) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_MOUSE
|
||||||
| NC score | 0.257145 (rank : 25) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.255023 (rank : 26) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.252904 (rank : 27) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.251495 (rank : 28) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.237151 (rank : 29) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.230543 (rank : 30) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.193146 (rank : 31) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.192231 (rank : 32) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.095988 (rank : 33) | θ value | 0.000461057 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.093787 (rank : 34) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.092388 (rank : 35) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.089855 (rank : 36) | θ value | 0.00228821 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.054818 (rank : 37) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
SNX15_MOUSE
|
||||||
| NC score | 0.051628 (rank : 38) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.051231 (rank : 39) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.051231 (rank : 40) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.050271 (rank : 41) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.048044 (rank : 42) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.047817 (rank : 43) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.044712 (rank : 44) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.038343 (rank : 45) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
K0146_MOUSE
|
||||||
| NC score | 0.035217 (rank : 46) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
RT09_HUMAN
|
||||||
| NC score | 0.030006 (rank : 47) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82933 | Gene names | MRPS9, RPMS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S9, mitochondrial precursor (S9mt) (MRP-S9). | |||||
|
VPS18_HUMAN
|
||||||
| NC score | 0.029131 (rank : 48) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
KS6C1_HUMAN
|
||||||
| NC score | 0.027220 (rank : 49) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
VPS18_MOUSE
|
||||||
| NC score | 0.026957 (rank : 50) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
KS6C1_MOUSE
|
||||||
| NC score | 0.024532 (rank : 51) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.023589 (rank : 52) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.023450 (rank : 53) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TPSNR_MOUSE
|
||||||
| NC score | 0.020113 (rank : 54) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.018281 (rank : 55) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.018191 (rank : 56) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
RPKL1_HUMAN
|
||||||
| NC score | 0.016674 (rank : 57) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6S9, Q69YT9, Q6ZMQ6, Q96NR9, Q9BSU9 | Gene names | RPS6KL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.016667 (rank : 58) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.016495 (rank : 59) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.014163 (rank : 60) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
FBX9_HUMAN
|
||||||
| NC score | 0.013469 (rank : 61) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK97, O75986, Q6PKH7, Q9NT57, Q9Y593 | Gene names | FBXO9, FBX9, VCIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 9 (NY-REN-57 antigen) (Cross-immune reaction antigen 1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.013336 (rank : 62) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.012635 (rank : 63) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.012609 (rank : 64) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.011962 (rank : 65) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
ARI1_HUMAN
|
||||||
| NC score | 0.011812 (rank : 66) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| NC score | 0.011807 (rank : 67) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.011734 (rank : 68) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.011597 (rank : 69) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.011506 (rank : 70) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.010735 (rank : 71) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.010698 (rank : 72) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.010585 (rank : 73) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.010495 (rank : 74) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.009371 (rank : 75) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ULK3_MOUSE
|
||||||
| NC score | 0.009282 (rank : 76) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3U3Q1, Q8K1X6, Q9DBR8 | Gene names | Ulk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK3 (EC 2.7.11.1) (Unc-51-like kinase 3). | |||||
|
FLNB_MOUSE
|
||||||
| NC score | 0.009031 (rank : 77) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
CERU_MOUSE
|
||||||
| NC score | 0.008422 (rank : 78) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.008391 (rank : 79) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.008031 (rank : 80) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.007863 (rank : 81) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
FBLN3_MOUSE
|
||||||
| NC score | 0.006577 (rank : 82) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.005793 (rank : 83) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
FBLN3_HUMAN
|
||||||
| NC score | 0.004494 (rank : 84) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||