Please be patient as the page loads
|
PYR1_HUMAN
|
||||||
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CPSM_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.918234 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.917954 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
OTC_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 4) | NC score | 0.364949 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11725 | Gene names | Otc | |||
|
Domain Architecture |
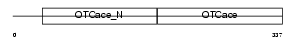
|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
OTC_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 5) | NC score | 0.360353 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00480, Q6B0I1, Q9NYJ5 | Gene names | OTC | |||
|
Domain Architecture |
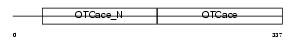
|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 6) | NC score | 0.408544 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 7) | NC score | 0.403585 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 8) | NC score | 0.396358 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 9) | NC score | 0.351412 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 10) | NC score | 0.392575 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 11) | NC score | 0.393927 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
DPYL4_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 12) | NC score | 0.259396 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35098, O08886 | Gene names | Dpysl4, Crmp3, Ulip4 | |||
|
Domain Architecture |
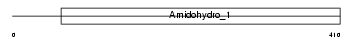
|
|||||
| Description | Dihydropyrimidinase-related protein 4 (DRP-4) (Collapsin response mediator protein 3) (CRMP-3) (UNC33-like phosphoprotein 4) (ULIP4 protein). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 13) | NC score | 0.391790 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
DPYL4_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 14) | NC score | 0.255802 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14531, O00240, Q5T0Q7 | Gene names | DPYSL4, CRMP3, ULIP4 | |||
|
Domain Architecture |
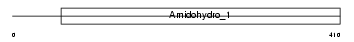
|
|||||
| Description | Dihydropyrimidinase-related protein 4 (DRP-4) (Collapsin response mediator protein 3) (CRMP-3) (UNC33-like phosphoprotein 4) (ULIP4 protein). | |||||
|
DPYL5_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 15) | NC score | 0.253278 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQF6 | Gene names | Dpysl5, Crmp5 | |||
|
Domain Architecture |
|
|||||
| Description | Dihydropyrimidinase-related protein 5 (DRP-5) (Collapsin response mediator protein 5) (CRMP-5). | |||||
|
DPYL3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.247706 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14195, Q93012 | Gene names | DPYSL3, CRMP4, DRP3, ULIP | |||
|
Domain Architecture |
|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein) (Collapsin response mediator protein 4) (CRMP-4). | |||||
|
DPYL3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 17) | NC score | 0.247768 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |
|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
DPYL5_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 18) | NC score | 0.248148 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BPU6, Q8TCL6, Q9NQC4, Q9NRY9 | Gene names | DPYSL5, CRMP5, ULIP6 | |||
|
Domain Architecture |
|
|||||
| Description | Dihydropyrimidinase-related protein 5 (DRP-5) (ULIP6 protein) (Collapsin response mediator protein 5) (CRMP-5) (CRMP3-associated molecule) (CRAM). | |||||
|
DPYS_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 19) | NC score | 0.246805 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQF5, Q99PP1, Q9DBK3, Q9DBP7 | Gene names | Dpys | |||
|
Domain Architecture |
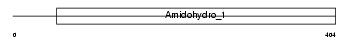
|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
DPYL2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 20) | NC score | 0.245323 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16555, O00424 | Gene names | DPYSL2, CRMP2 | |||
|
Domain Architecture |
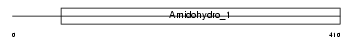
|
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (Collapsin response mediator protein 2) (CRMP-2) (N2A3). | |||||
|
DPYL2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 21) | NC score | 0.244051 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08553, Q6P5D0 | Gene names | Dpysl2, Crmp2, Ulip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (ULIP 2 protein). | |||||
|
DPYL1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 22) | NC score | 0.243651 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97427, O08554, O35097 | Gene names | Crmp1, Dpysl1, Ulip3 | |||
|
Domain Architecture |
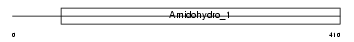
|
|||||
| Description | Dihydropyrimidinase-related protein 1 (DRP-1) (Collapsin response mediator protein 1) (CRMP-1) (ULIP3 protein). | |||||
|
DPYS_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 23) | NC score | 0.244718 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14117 | Gene names | DPYS | |||
|
Domain Architecture |
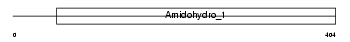
|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 24) | NC score | 0.367421 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
DPYL1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 25) | NC score | 0.241371 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14194, Q13024, Q96TC8 | Gene names | CRMP1, DPYSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 1 (DRP-1) (Collapsin response mediator protein 1) (CRMP-1). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.067833 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
EGF_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 27) | NC score | 0.022308 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.009828 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
ADA2B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 29) | NC score | 0.007540 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||
|
GUAA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.142600 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49915 | Gene names | GMPS | |||
|
Domain Architecture |

|
|||||
| Description | GMP synthase [glutamine-hydrolyzing] (EC 6.3.5.2) (Glutamine amidotransferase) (GMP synthetase). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.023808 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
PUR2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.085450 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
LCP2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.049573 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.030014 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.041053 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
NGN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.026204 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.025292 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.014228 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
CS021_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.034481 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.012159 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.036365 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.008085 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.043103 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.018415 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TNK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.006967 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.039557 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.039717 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
FOXI2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.008929 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3I5G5, Q8BIK9 | Gene names | Foxi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I2. | |||||
|
HXA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.005481 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.028333 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
DYN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.014987 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
INGR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.027946 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15260 | Gene names | IFNGR1 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen) (CDw119). | |||||
|
M3K1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.002146 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||
|
NETR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.004045 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.012281 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.019942 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.019901 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.009319 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.023951 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.014967 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.020328 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.022350 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF1C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.023983 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.031478 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.011460 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.011607 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.000446 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.023334 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.031795 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
STRN3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.012702 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERG2 | Gene names | Strn3, Gs2na, Sg2na | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-3 (Cell-cycle autoantigen SG2NA) (S/G2 antigen). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.021082 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.022124 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.006171 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
FAK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.002603 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P34152, O08578 | Gene names | Ptk2, Fadk, Fak, Fak1 | |||
|
Domain Architecture |

|
|||||
| Description | Focal adhesion kinase 1 (EC 2.7.10.2) (FADK 1) (pp125FAK). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.015939 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HIC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.000107 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.017741 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.021465 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.008790 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.004163 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.010477 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.007408 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CO8A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.011727 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25067, Q8TEJ5 | Gene names | COL8A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
CO8A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.012121 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
CU086_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.028993 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.009993 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.017640 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
IER5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.019817 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
LNP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.018095 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.007592 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.004537 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.015345 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.014519 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
STRN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.011391 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRL3 | Gene names | STRN4, ZIN | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-4 (Zinedin). | |||||
|
STRN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.011446 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58404, Q68EF5 | Gene names | Strn4, Zin | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-4 (Zinedin). | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | -0.000348 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.023222 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.014262 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.014264 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
EGF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.012803 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.007472 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.006627 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.008814 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.006376 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
SCOT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.011926 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYC2, Q8NHR1, Q9H1I4 | Gene names | OXCT2 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA:3-ketoacid-coenzyme A transferase 2, mitochondrial precursor (EC 2.8.3.5) (Testis-specific succinyl CoA:3-oxoacid CoA- transferase) (SCOT-t). | |||||
|
SPTA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.025945 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9T6, Q80ZS4, Q9D5L5, Q9DAF5 | Gene names | Spata3, Tsarg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1) (mTSARG1). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.019727 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.018086 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009117 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
E2F3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.006957 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
GLPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.017687 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |
|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.015491 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
RSMN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.013873 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.013873 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.000005 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.000742 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TNK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.004581 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
VINC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.011767 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |
|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
VINC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010712 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
PUR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.070447 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.918234 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.917954 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.408544 (rank : 4) | θ value | 1.2105e-12 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.403585 (rank : 5) | θ value | 4.59992e-12 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.396358 (rank : 6) | θ value | 7.84624e-12 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.393927 (rank : 7) | θ value | 1.63604e-09 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.392575 (rank : 8) | θ value | 9.59137e-10 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.391790 (rank : 9) | θ value | 1.99992e-07 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.367421 (rank : 10) | θ value | 0.00035302 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
OTC_MOUSE
|
||||||
| NC score | 0.364949 (rank : 11) | θ value | 2.06002e-20 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11725 | Gene names | Otc | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
OTC_HUMAN
|
||||||
| NC score | 0.360353 (rank : 12) | θ value | 1.33526e-19 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00480, Q6B0I1, Q9NYJ5 | Gene names | OTC | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.351412 (rank : 13) | θ value | 7.34386e-10 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
DPYL4_MOUSE
|
||||||
| NC score | 0.259396 (rank : 14) | θ value | 1.80886e-08 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35098, O08886 | Gene names | Dpysl4, Crmp3, Ulip4 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 4 (DRP-4) (Collapsin response mediator protein 3) (CRMP-3) (UNC33-like phosphoprotein 4) (ULIP4 protein). | |||||
|
DPYL4_HUMAN
|
||||||
| NC score | 0.255802 (rank : 15) | θ value | 2.61198e-07 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14531, O00240, Q5T0Q7 | Gene names | DPYSL4, CRMP3, ULIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 4 (DRP-4) (Collapsin response mediator protein 3) (CRMP-3) (UNC33-like phosphoprotein 4) (ULIP4 protein). | |||||
|
DPYL5_MOUSE
|
||||||
| NC score | 0.253278 (rank : 16) | θ value | 2.88788e-06 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQF6 | Gene names | Dpysl5, Crmp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 5 (DRP-5) (Collapsin response mediator protein 5) (CRMP-5). | |||||
|
DPYL5_HUMAN
|
||||||
| NC score | 0.248148 (rank : 17) | θ value | 7.1131e-05 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BPU6, Q8TCL6, Q9NQC4, Q9NRY9 | Gene names | DPYSL5, CRMP5, ULIP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 5 (DRP-5) (ULIP6 protein) (Collapsin response mediator protein 5) (CRMP-5) (CRMP3-associated molecule) (CRAM). | |||||
|
DPYL3_MOUSE
|
||||||
| NC score | 0.247768 (rank : 18) | θ value | 3.19293e-05 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
DPYL3_HUMAN
|
||||||
| NC score | 0.247706 (rank : 19) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14195, Q93012 | Gene names | DPYSL3, CRMP4, DRP3, ULIP | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein) (Collapsin response mediator protein 4) (CRMP-4). | |||||
|
DPYS_MOUSE
|
||||||
| NC score | 0.246805 (rank : 20) | θ value | 9.29e-05 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQF5, Q99PP1, Q9DBK3, Q9DBP7 | Gene names | Dpys | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
DPYL2_HUMAN
|
||||||
| NC score | 0.245323 (rank : 21) | θ value | 0.000121331 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16555, O00424 | Gene names | DPYSL2, CRMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (Collapsin response mediator protein 2) (CRMP-2) (N2A3). | |||||
|
DPYS_HUMAN
|
||||||
| NC score | 0.244718 (rank : 22) | θ value | 0.00035302 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14117 | Gene names | DPYS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
DPYL2_MOUSE
|
||||||
| NC score | 0.244051 (rank : 23) | θ value | 0.000270298 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08553, Q6P5D0 | Gene names | Dpysl2, Crmp2, Ulip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (ULIP 2 protein). | |||||
|
DPYL1_MOUSE
|
||||||
| NC score | 0.243651 (rank : 24) | θ value | 0.00035302 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97427, O08554, O35097 | Gene names | Crmp1, Dpysl1, Ulip3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 1 (DRP-1) (Collapsin response mediator protein 1) (CRMP-1) (ULIP3 protein). | |||||
|
DPYL1_HUMAN
|
||||||
| NC score | 0.241371 (rank : 25) | θ value | 0.00134147 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14194, Q13024, Q96TC8 | Gene names | CRMP1, DPYSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 1 (DRP-1) (Collapsin response mediator protein 1) (CRMP-1). | |||||
|
GUAA_HUMAN
|
||||||
| NC score | 0.142600 (rank : 26) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49915 | Gene names | GMPS | |||
|
Domain Architecture |

|
|||||
| Description | GMP synthase [glutamine-hydrolyzing] (EC 6.3.5.2) (Glutamine amidotransferase) (GMP synthetase). | |||||
|
PUR2_HUMAN
|
||||||
| NC score | 0.085450 (rank : 27) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
PUR2_MOUSE
|
||||||
| NC score | 0.070447 (rank : 28) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.067833 (rank : 29) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
LCP2_MOUSE
|
||||||
| NC score | 0.049573 (rank : 30) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.043103 (rank : 31) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.041053 (rank : 32) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.039717 (rank : 33) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.039557 (rank : 34) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.036365 (rank : 35) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CS021_MOUSE
|
||||||
| NC score | 0.034481 (rank : 36) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.031795 (rank : 37) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.031478 (rank : 38) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.030014 (rank : 39) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
CU086_HUMAN
|
||||||
| NC score | 0.028993 (rank : 40) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.028333 (rank : 41) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
INGR1_HUMAN
|
||||||
| NC score | 0.027946 (rank : 42) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15260 | Gene names | IFNGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen) (CDw119). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.026204 (rank : 43) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
SPTA3_MOUSE
|
||||||
| NC score | 0.025945 (rank : 44) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9T6, Q80ZS4, Q9D5L5, Q9DAF5 | Gene names | Spata3, Tsarg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1) (mTSARG1). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.025292 (rank : 45) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
TAF1C_HUMAN
|
||||||
| NC score | 0.023983 (rank : 46) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.023951 (rank : 47) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.023808 (rank : 48) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.023334 (rank : 49) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.023222 (rank : 50) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.022350 (rank : 51) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.022308 (rank : 52) | θ value | 0.0148317 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.022124 (rank : 53) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.021465 (rank : 54) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.021082 (rank : 55) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.020328 (rank : 56) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.019942 (rank : 57) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.019901 (rank : 58) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
IER5_MOUSE
|
||||||
| NC score | 0.019817 (rank : 59) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.019727 (rank : 60) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.018415 (rank : 61) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
LNP_MOUSE
|
||||||
| NC score | 0.018095 (rank : 62) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.018086 (rank : 63) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.017741 (rank : 64) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
GLPC_HUMAN
|
||||||
| NC score | 0.017687 (rank : 65) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.017640 (rank : 66) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.015939 (rank : 67) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.015491 (rank : 68) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.015345 (rank : 69) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.014987 (rank : 70) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.014967 (rank : 71) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.014519 (rank : 72) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.014264 (rank : 73) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.014262 (rank : 74) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.014228 (rank : 75) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.013873 (rank : 76) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.013873 (rank : 77) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
EGF_HUMAN
|
||||||
| NC score | 0.012803 (rank : 78) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
STRN3_MOUSE
|
||||||
| NC score | 0.012702 (rank : 79) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERG2 | Gene names | Strn3, Gs2na, Sg2na | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-3 (Cell-cycle autoantigen SG2NA) (S/G2 antigen). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.012281 (rank : 80) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.012159 (rank : 81) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
CO8A2_MOUSE
|
||||||
| NC score | 0.012121 (rank : 82) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
SCOT2_HUMAN
|
||||||
| NC score | 0.011926 (rank : 83) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYC2, Q8NHR1, Q9H1I4 | Gene names | OXCT2 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA:3-ketoacid-coenzyme A transferase 2, mitochondrial precursor (EC 2.8.3.5) (Testis-specific succinyl CoA:3-oxoacid CoA- transferase) (SCOT-t). | |||||
|
VINC_HUMAN
|
||||||
| NC score | 0.011767 (rank : 84) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
CO8A2_HUMAN
|
||||||
| NC score | 0.011727 (rank : 85) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25067, Q8TEJ5 | Gene names | COL8A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.011607 (rank : 86) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.011460 (rank : 87) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
STRN4_MOUSE
|
||||||
| NC score | 0.011446 (rank : 88) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58404, Q68EF5 | Gene names | Strn4, Zin | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-4 (Zinedin). | |||||
|
STRN4_HUMAN
|
||||||
| NC score | 0.011391 (rank : 89) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRL3 | Gene names | STRN4, ZIN | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-4 (Zinedin). | |||||
|
VINC_MOUSE
|
||||||
| NC score | 0.010712 (rank : 90) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.010477 (rank : 91) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.009993 (rank : 92) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.009828 (rank : 93) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.009319 (rank : 94) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.009117 (rank : 95) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
FOXI2_MOUSE
|
||||||
| NC score | 0.008929 (rank : 96) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3I5G5, Q8BIK9 | Gene names | Foxi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I2. | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.008814 (rank : 97) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.008790 (rank : 98) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.008085 (rank : 99) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.007592 (rank : 100) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
ADA2B_MOUSE
|
||||||
| NC score | 0.007540 (rank : 101) | θ value | 0.0563607 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.007472 (rank : 102) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.007408 (rank : 103) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
TNK1_HUMAN
|
||||||
| NC score | 0.006967 (rank : 104) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
E2F3_MOUSE
|
||||||
| NC score | 0.006957 (rank : 105) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.006627 (rank : 106) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.006376 (rank : 107) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.006171 (rank : 108) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
HXA4_MOUSE
|
||||||
| NC score | 0.005481 (rank : 109) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
TNK1_MOUSE
|
||||||
| NC score | 0.004581 (rank : 110) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99ML2, Q8CCC4, Q8K0X9, Q91V11 | Gene names | Tnk1, Kos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (Kinase of embryonic stem cells). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.004537 (rank : 111) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.004163 (rank : 112) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.004045 (rank : 113) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
FAK1_MOUSE
|
||||||
| NC score | 0.002603 (rank : 114) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P34152, O08578 | Gene names | Ptk2, Fadk, Fak, Fak1 | |||
|
Domain Architecture |

|
|||||
| Description | Focal adhesion kinase 1 (EC 2.7.10.2) (FADK 1) (pp125FAK). | |||||
|
M3K1_MOUSE
|
||||||
| NC score | 0.002146 (rank : 115) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.000742 (rank : 116) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
HIC1_MOUSE
|
||||||
| NC score | 0.000446 (rank : 117) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
HIC1_HUMAN
|
||||||
| NC score | 0.000107 (rank : 118) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.000005 (rank : 119) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ZN692_MOUSE
|
||||||
| NC score | -0.000348 (rank : 120) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||