Please be patient as the page loads
|
DYN2_HUMAN
|
||||||
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DYN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979150 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981951 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996797 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.985646 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DNM1L_HUMAN
|
||||||
| θ value | 6.21357e-126 (rank : 6) | NC score | 0.947469 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
DNM1L_MOUSE
|
||||||
| θ value | 1.38424e-125 (rank : 7) | NC score | 0.947736 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
MX1_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 8) | NC score | 0.866177 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| θ value | 3.14151e-61 (rank : 9) | NC score | 0.864745 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 10) | NC score | 0.864274 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 11) | NC score | 0.858573 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 12) | NC score | 0.690223 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 13) | NC score | 0.693121 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 14) | NC score | 0.156055 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 15) | NC score | 0.152836 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 16) | NC score | 0.074522 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.158218 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
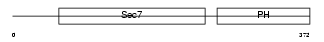
CYH1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.158130 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.016848 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.157017 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.157120 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.149607 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.149575 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 24) | NC score | 0.138656 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.129629 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.038201 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 27) | NC score | 0.124925 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 28) | NC score | 0.090986 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.092505 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 30) | NC score | 0.132628 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
GAK_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.012195 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 32) | NC score | 0.070812 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.127881 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.047982 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
K2C5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.023422 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.063139 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.032697 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.018062 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.072667 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.078423 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.027073 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.022969 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.016545 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.021759 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.077829 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.083463 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.082373 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.086463 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.019466 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
K2C5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.019655 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.040580 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.013450 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.169156 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.014918 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.020080 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.104386 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.100218 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.086101 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
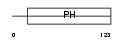
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.011683 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.011274 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.007647 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.014873 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.017333 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.020125 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
K2C6B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.020651 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.014767 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
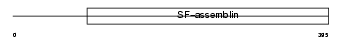
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.021456 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PKHB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.078993 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |
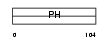
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.014987 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.023262 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.051675 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.031426 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.030496 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.029615 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.022288 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.033134 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.006373 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.028077 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.041384 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.018331 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.074897 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.005273 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.010597 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.027460 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
K2C6A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.016616 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
K2C6C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.016605 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
UFO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | -0.001027 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30530 | Gene names | AXL, UFO | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (AXL oncogene). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.013953 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.015134 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
NFIC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.009841 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |
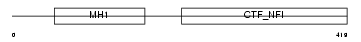
|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.006746 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
SON_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.029076 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.004981 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.012416 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
UGGG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.021807 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | -0.000220 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.025577 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.021044 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.018547 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.022309 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.008972 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.011109 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.001982 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
GIMA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.009144 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUV9 | Gene names | GIMAP4, IAN1, IMAP4 | |||
|
Domain Architecture |
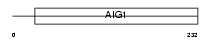
|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein) (hIAN1). | |||||
|
K2C6A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.015044 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.006430 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.023178 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.034737 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PICAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.008516 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
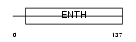
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.036630 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TBX6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.002304 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |
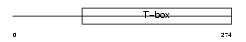
|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.016512 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
BIG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.052141 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
BIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052593 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
GBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.056489 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050587 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051315 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PSD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.058312 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
PSD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052438 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| NC score | 0.996797 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.985646 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.981951 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.979150 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DNM1L_MOUSE
|
||||||
| NC score | 0.947736 (rank : 6) | θ value | 1.38424e-125 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
DNM1L_HUMAN
|
||||||
| NC score | 0.947469 (rank : 7) | θ value | 6.21357e-126 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
MX1_HUMAN
|
||||||
| NC score | 0.866177 (rank : 8) | θ value | 9.76566e-63 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| NC score | 0.864745 (rank : 9) | θ value | 3.14151e-61 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| NC score | 0.864274 (rank : 10) | θ value | 1.7238e-59 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| NC score | 0.858573 (rank : 11) | θ value | 2.48917e-58 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_MOUSE
|
||||||
| NC score | 0.693121 (rank : 12) | θ value | 3.07116e-24 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.690223 (rank : 13) | θ value | 3.07116e-24 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.169156 (rank : 14) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.158218 (rank : 15) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.158130 (rank : 16) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.157120 (rank : 17) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.157017 (rank : 18) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.156055 (rank : 19) | θ value | 0.000602161 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.152836 (rank : 20) | θ value | 0.000602161 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.149607 (rank : 21) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.149575 (rank : 22) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.138656 (rank : 23) | θ value | 0.00869519 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.132628 (rank : 24) | θ value | 0.0330416 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.129629 (rank : 25) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.127881 (rank : 26) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.124925 (rank : 27) | θ value | 0.0252991 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.104386 (rank : 28) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.100218 (rank : 29) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.092505 (rank : 30) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.090986 (rank : 31) | θ value | 0.0252991 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.086463 (rank : 32) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.086101 (rank : 33) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.083463 (rank : 34) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.082373 (rank : 35) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHB1_MOUSE
|
||||||
| NC score | 0.078993 (rank : 36) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.078423 (rank : 37) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.077829 (rank : 38) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.074897 (rank : 39) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.074522 (rank : 40) | θ value | 0.000602161 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.072667 (rank : 41) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.070812 (rank : 42) | θ value | 0.0563607 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.063139 (rank : 43) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PSD3_HUMAN
|
||||||
| NC score | 0.058312 (rank : 44) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.056489 (rank : 45) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.052593 (rank : 46) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.052438 (rank : 47) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
BIG1_HUMAN
|
||||||
| NC score | 0.052141 (rank : 48) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.051675 (rank : 49) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.051315 (rank : 50) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.050587 (rank : 51) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.047982 (rank : 52) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.041384 (rank : 53) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.040580 (rank : 54) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.038201 (rank : 55) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.036630 (rank : 56) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.034737 (rank : 57) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.033134 (rank : 58) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.032697 (rank : 59) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.031426 (rank : 60) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.030496 (rank : 61) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.029615 (rank : 62) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.029076 (rank : 63) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.028077 (rank : 64) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.027460 (rank : 65) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.027073 (rank : 66) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.025577 (rank : 67) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
K2C5_MOUSE
|
||||||
| NC score | 0.023422 (rank : 68) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.023262 (rank : 69) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.023178 (rank : 70) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.022969 (rank : 71) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.022309 (rank : 72) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.022288 (rank : 73) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
UGGG1_MOUSE
|
||||||
| NC score | 0.021807 (rank : 74) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.021759 (rank : 75) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.021456 (rank : 76) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.021044 (rank : 77) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
K2C6B_MOUSE
|
||||||
| NC score | 0.020651 (rank : 78) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.020125 (rank : 79) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.020080 (rank : 80) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
K2C5_HUMAN
|
||||||
| NC score | 0.019655 (rank : 81) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.019466 (rank : 82) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.018547 (rank : 83) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.018331 (rank : 84) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.018062 (rank : 85) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.017333 (rank : 86) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.016848 (rank : 87) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
K2C6A_HUMAN
|
||||||
| NC score | 0.016616 (rank : 88) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
K2C6C_HUMAN
|
||||||
| NC score | 0.016605 (rank : 89) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.016545 (rank : 90) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.016512 (rank : 91) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.015134 (rank : 92) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
K2C6A_MOUSE
|
||||||
| NC score | 0.015044 (rank : 93) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.014987 (rank : 94) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.014918 (rank : 95) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.014873 (rank : 96) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.014767 (rank : 97) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.013953 (rank : 98) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.013450 (rank : 99) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.012416 (rank : 100) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.012195 (rank : 101) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.011683 (rank : 102) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.011274 (rank : 103) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.011109 (rank : 104) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.010597 (rank : 105) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
NFIC_MOUSE
|
||||||
| NC score | 0.009841 (rank : 106) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
GIMA4_HUMAN
|
||||||
| NC score | 0.009144 (rank : 107) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUV9 | Gene names | GIMAP4, IAN1, IMAP4 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase, IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein) (hIAN1). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.008972 (rank : 108) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.008516 (rank : 109) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.007647 (rank : 110) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.006746 (rank : 111) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.006430 (rank : 112) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.006373 (rank : 113) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.005273 (rank : 114) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.004981 (rank : 115) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TBX6_MOUSE
|
||||||
| NC score | 0.002304 (rank : 116) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.001982 (rank : 117) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.000220 (rank : 118) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
UFO_HUMAN
|
||||||
| NC score | -0.001027 (rank : 119) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30530 | Gene names | AXL, UFO | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (AXL oncogene). | |||||