Please be patient as the page loads
|
DYN3_HUMAN
|
||||||
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DYN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977281 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980064 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.985646 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.982752 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DNM1L_HUMAN
|
||||||
| θ value | 1.0968e-122 (rank : 6) | NC score | 0.941171 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
DNM1L_MOUSE
|
||||||
| θ value | 4.16784e-122 (rank : 7) | NC score | 0.941301 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
MX1_HUMAN
|
||||||
| θ value | 3.47335e-60 (rank : 8) | NC score | 0.859332 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| θ value | 1.11734e-58 (rank : 9) | NC score | 0.858062 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| θ value | 6.13105e-57 (rank : 10) | NC score | 0.857464 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| θ value | 6.13105e-57 (rank : 11) | NC score | 0.851947 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 12) | NC score | 0.679767 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 13) | NC score | 0.682724 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 14) | NC score | 0.081242 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.088422 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 16) | NC score | 0.046854 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.155719 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.158487 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 19) | NC score | 0.158289 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
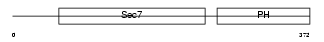
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 20) | NC score | 0.149940 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 21) | NC score | 0.149911 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.152798 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
K2C4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 23) | NC score | 0.010728 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 24) | NC score | 0.055008 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.076665 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 26) | NC score | 0.173514 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 27) | NC score | 0.081827 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.133188 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 29) | NC score | 0.031200 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.036709 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 31) | NC score | 0.126767 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.128241 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.087477 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.021083 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.145996 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.146103 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.121248 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
LATS2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.006275 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.026317 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.115919 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.091786 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
HXB13_MOUSE
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.008561 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70321, Q80Y12 | Gene names | Hoxb13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.029620 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.014386 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.060879 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
RNF25_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.036311 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |
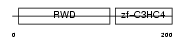
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
UGGG1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.039367 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.039951 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.055950 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.083325 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.084879 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.053421 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.079718 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TS13_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.036356 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.006948 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.001583 (rank : 191) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.009707 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.045613 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.043829 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.010704 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
UGGG1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.030184 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.082547 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.079648 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.034454 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.065927 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.006066 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.039254 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.038166 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.026212 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.027025 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.014743 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.032395 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.040496 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CU086_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.049758 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.038581 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.036185 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.043960 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.026230 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.039469 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.036412 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.006425 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.031499 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.013681 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.041090 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.010130 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.012335 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.018322 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.026590 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.015003 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.012873 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
GCM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.014190 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |
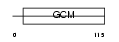
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.015870 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.027618 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.072961 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.071382 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.006871 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.096481 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PPIP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.012771 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M15, Q9Z189 | Gene names | Pstpip2, Mayp | |||
|
Domain Architecture |
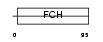
|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2 (Macrophage actin-associated tyrosine-phosphorylated protein) (pp37). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.012276 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.015026 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.039078 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.034010 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.017185 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.020184 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
HERPU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.015964 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15011, O60644, Q96D92 | Gene names | HERPUD1, KIAA0025, MIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein (Methyl methanesulfonate (MMF)-inducible fragment protein 1). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.020814 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.027143 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
OAS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.010995 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.009209 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.016638 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.039484 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RNF25_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.026505 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |
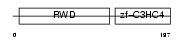
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.023391 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.027734 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.002774 (rank : 188) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
GP153_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.008322 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0Z9, Q80T37, Q8CFK0 | Gene names | Gpr153, Pgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 153 (G-protein coupled receptor PGR1). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.021359 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.026316 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.006760 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.024894 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.025961 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.007072 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.044339 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.006218 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.035624 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.035710 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.009402 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.053730 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.008964 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.016641 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.010473 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
APOL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.010439 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWW9, Q5TFL9, Q9UGW5 | Gene names | APOL5 | |||
|
Domain Architecture |
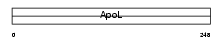
|
|||||
| Description | Apolipoprotein-L5 (Apolipoprotein L-V) (ApoL-V). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.015991 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.029748 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.010704 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.026619 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.010209 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.044025 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.029857 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.003930 (rank : 187) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.017935 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.054166 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.014243 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.010733 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.006594 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.040986 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.020672 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.006781 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.009713 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.002553 (rank : 189) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.002531 (rank : 190) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.054430 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PICAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.011376 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
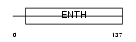
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
PICA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.008565 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |
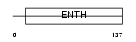
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.017738 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.011845 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.016904 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TNR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.004831 (rank : 186) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
BIG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.052616 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
BIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.053145 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.057284 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.061035 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.057618 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
GBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.058854 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053426 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.056738 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.057042 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052446 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.052633 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.061377 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.089957 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.067346 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PKHB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.060608 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.063756 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.064002 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.066455 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.072634 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059932 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.053101 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PSD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.057783 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
PSD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.056115 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.054011 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055103 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052313 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.050629 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.064047 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.061281 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.051341 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.050346 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.051839 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050278 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.985646 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| NC score | 0.982752 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.980064 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.977281 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DNM1L_MOUSE
|
||||||
| NC score | 0.941301 (rank : 6) | θ value | 4.16784e-122 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
DNM1L_HUMAN
|
||||||
| NC score | 0.941171 (rank : 7) | θ value | 1.0968e-122 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
MX1_HUMAN
|
||||||
| NC score | 0.859332 (rank : 8) | θ value | 3.47335e-60 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| NC score | 0.858062 (rank : 9) | θ value | 1.11734e-58 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| NC score | 0.857464 (rank : 10) | θ value | 6.13105e-57 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| NC score | 0.851947 (rank : 11) | θ value | 6.13105e-57 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_MOUSE
|
||||||
| NC score | 0.682724 (rank : 12) | θ value | 1.86321e-21 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.679767 (rank : 13) | θ value | 1.86321e-21 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.173514 (rank : 14) | θ value | 0.0113563 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.158487 (rank : 15) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.158289 (rank : 16) | θ value | 0.00134147 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.155719 (rank : 17) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.152798 (rank : 18) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.149940 (rank : 19) | θ value | 0.00175202 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.149911 (rank : 20) | θ value | 0.00175202 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.146103 (rank : 21) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.145996 (rank : 22) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.133188 (rank : 23) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.128241 (rank : 24) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.126767 (rank : 25) | θ value | 0.0431538 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.121248 (rank : 26) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.115919 (rank : 27) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.096481 (rank : 28) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.091786 (rank : 29) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.089957 (rank : 30) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.088422 (rank : 31) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.087477 (rank : 32) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.084879 (rank : 33) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.083325 (rank : 34) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.082547 (rank : 35) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.081827 (rank : 36) | θ value | 0.0148317 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.081242 (rank : 37) | θ value | 2.61198e-07 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.079718 (rank : 38) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.079648 (rank : 39) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.076665 (rank : 40) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.072961 (rank : 41) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.072634 (rank : 42) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.071382 (rank : 43) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.067346 (rank : 44) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.066455 (rank : 45) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.065927 (rank : 46) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.064047 (rank : 47) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.064002 (rank : 48) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.063756 (rank : 49) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.061377 (rank : 50) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.061281 (rank : 51) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.061035 (rank : 52) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.060879 (rank : 53) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PKHB1_MOUSE
|
||||||
| NC score | 0.060608 (rank : 54) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.059932 (rank : 55) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.058854 (rank : 56) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
PSD3_HUMAN
|
||||||
| NC score | 0.057783 (rank : 57) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.057618 (rank : 58) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.057284 (rank : 59) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.057042 (rank : 60) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.056738 (rank : 61) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.056115 (rank : 62) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.055950 (rank : 63) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.055103 (rank : 64) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.055008 (rank : 65) | θ value | 0.00869519 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.054430 (rank : 66) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.054166 (rank : 67) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.054011 (rank : 68) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.053730 (rank : 69) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.053426 (rank : 70) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.053421 (rank : 71) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.053145 (rank : 72) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.053101 (rank : 73) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.052633 (rank : 74) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
BIG1_HUMAN
|
||||||
| NC score | 0.052616 (rank : 75) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.052446 (rank : 76) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.052313 (rank : 77) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.051839 (rank : 78) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.051341 (rank : 79) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.050629 (rank : 80) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.050346 (rank : 81) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.050278 (rank : 82) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
CU086_HUMAN
|
||||||
| NC score | 0.049758 (rank : 83) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.046854 (rank : 84) | θ value | 0.000602161 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.045613 (rank : 85) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.044339 (rank : 86) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.044025 (rank : 87) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.043960 (rank : 88) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.043829 (rank : 89) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.041090 (rank : 90) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.040986 (rank : 91) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.040496 (rank : 92) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.039951 (rank : 93) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.039484 (rank : 94) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.039469 (rank : 95) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
UGGG1_MOUSE
|
||||||
| NC score | 0.039367 (rank : 96) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.039254 (rank : 97) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.039078 (rank : 98) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.038581 (rank : 99) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.038166 (rank : 100) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.036709 (rank : 101) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.036412 (rank : 102) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.036356 (rank : 103) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
RNF25_HUMAN
|
||||||
| NC score | 0.036311 (rank : 104) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.036185 (rank : 105) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.035710 (rank : 106) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.035624 (rank : 107) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.034454 (rank : 108) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.034010 (rank : 109) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.032395 (rank : 110) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.031499 (rank : 111) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.031200 (rank : 112) | θ value | 0.0330416 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
UGGG1_HUMAN
|
||||||
| NC score | 0.030184 (rank : 113) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.029857 (rank : 114) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.029748 (rank : 115) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.029620 (rank : 116) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.027734 (rank : 117) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.027618 (rank : 118) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.027143 (rank : 119) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.027025 (rank : 120) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.026619 (rank : 121) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.026590 (rank : 122) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
RNF25_MOUSE
|
||||||
| NC score | 0.026505 (rank : 123) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.026317 (rank : 124) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.026316 (rank : 125) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.026230 (rank : 126) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.026212 (rank : 127) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.025961 (rank : 128) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.024894 (rank : 129) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.023391 (rank : 130) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.021359 (rank : 131) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.021083 (rank : 132) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.020814 (rank : 133) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.020672 (rank : 134) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.020184 (rank : 135) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.018322 (rank : 136) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.017935 (rank : 137) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.017738 (rank : 138) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.017185 (rank : 139) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.016904 (rank : 140) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.016641 (rank : 141) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.016638 (rank : 142) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.015991 (rank : 143) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
HERPU_HUMAN
|
||||||
| NC score | 0.015964 (rank : 144) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15011, O60644, Q96D92 | Gene names | HERPUD1, KIAA0025, MIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein (Methyl methanesulfonate (MMF)-inducible fragment protein 1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.015870 (rank : 145) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.015026 (rank : 146) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.015003 (rank : 147) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.014743 (rank : 148) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.014386 (rank : 149) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.014243 (rank : 150) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
GCM1_MOUSE
|
||||||
| NC score | 0.014190 (rank : 151) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.013681 (rank : 152) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.012873 (rank : 153) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
PPIP2_MOUSE
|
||||||
| NC score | 0.012771 (rank : 154) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M15, Q9Z189 | Gene names | Pstpip2, Mayp | |||
|
Domain Architecture |

|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2 (Macrophage actin-associated tyrosine-phosphorylated protein) (pp37). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.012335 (rank : 155) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.012276 (rank : 156) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.011845 (rank : 157) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.011376 (rank : 158) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
OAS3_HUMAN
|
||||||
| NC score | 0.010995 (rank : 159) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010733 (rank : 160) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
K2C4_HUMAN
|
||||||
| NC score | 0.010728 (rank : 161) | θ value | 0.00509761 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.010704 (rank : 162) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.010704 (rank : 163) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.010473 (rank : 164) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
APOL5_HUMAN
|
||||||
| NC score | 0.010439 (rank : 165) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWW9, Q5TFL9, Q9UGW5 | Gene names | APOL5 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein-L5 (Apolipoprotein L-V) (ApoL-V). | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.010209 (rank : 166) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.010130 (rank : 167) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.009713 (rank : 168) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.009707 (rank : 169) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.009402 (rank : 170) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.009209 (rank : 171) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.008964 (rank : 172) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
PICA_MOUSE
|
||||||
| NC score | 0.008565 (rank : 173) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
HXB13_MOUSE
|
||||||
| NC score | 0.008561 (rank : 174) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70321, Q80Y12 | Gene names | Hoxb13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
GP153_MOUSE
|
||||||
| NC score | 0.008322 (rank : 175) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0Z9, Q80T37, Q8CFK0 | Gene names | Gpr153, Pgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 153 (G-protein coupled receptor PGR1). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.007072 (rank : 176) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.006948 (rank : 177) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.006871 (rank : 178) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.006781 (rank : 179) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.006760 (rank : 180) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.006594 (rank : 181) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.006425 (rank : 182) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
LATS2_HUMAN
|
||||||
| NC score | 0.006275 (rank : 183) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.006218 (rank : 184) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.006066 (rank : 185) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
TNR3_HUMAN
|
||||||
| NC score | 0.004831 (rank : 186) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.003930 (rank : 187) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.002774 (rank : 188) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.002553 (rank : 189) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002531 (rank : 190) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.001583 (rank : 191) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||