Please be patient as the page loads
|
GCM1_MOUSE
|
||||||
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |
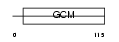
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987216 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |
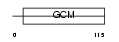
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
GCM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
GCM2_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 3) | NC score | 0.891417 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |
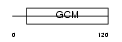
|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
GCM2_MOUSE
|
||||||
| θ value | 4.68348e-65 (rank : 4) | NC score | 0.895519 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09102, Q9QWX7, Q9Z289 | Gene names | Gcm2, Gcmb | |||
|
Domain Architecture |
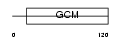
|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (mGCMb). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 5) | NC score | 0.039424 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.039189 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.048509 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.047078 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
NFIB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.020760 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.014190 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.029897 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.012892 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.028909 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.027722 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.028236 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
EGR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.001752 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.023395 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.016499 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.019220 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.015586 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.018497 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TR240_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.016607 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.005099 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GPV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.003789 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.004683 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.004954 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
C11B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.004985 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15539, Q64661 | Gene names | Cyp11b2, Cyp11b-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P450C11) (Steroid 11-beta-hydroxylase) (Aldosterone synthase). | |||||
|
DOCK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.007004 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
GCM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
GCM1_HUMAN
|
||||||
| NC score | 0.987216 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
GCM2_MOUSE
|
||||||
| NC score | 0.895519 (rank : 3) | θ value | 4.68348e-65 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09102, Q9QWX7, Q9Z289 | Gene names | Gcm2, Gcmb | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (mGCMb). | |||||
|
GCM2_HUMAN
|
||||||
| NC score | 0.891417 (rank : 4) | θ value | 2.4834e-66 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.048509 (rank : 5) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.047078 (rank : 6) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.039424 (rank : 7) | θ value | 0.365318 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.039189 (rank : 8) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.029897 (rank : 9) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.028909 (rank : 10) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.028236 (rank : 11) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.027722 (rank : 12) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.023395 (rank : 13) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NFIB_MOUSE
|
||||||
| NC score | 0.020760 (rank : 14) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.019220 (rank : 15) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.018497 (rank : 16) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TR240_HUMAN
|
||||||
| NC score | 0.016607 (rank : 17) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.016499 (rank : 18) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.015586 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.014190 (rank : 20) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.012892 (rank : 21) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
DOCK3_MOUSE
|
||||||
| NC score | 0.007004 (rank : 22) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.005099 (rank : 23) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
C11B2_MOUSE
|
||||||
| NC score | 0.004985 (rank : 24) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15539, Q64661 | Gene names | Cyp11b2, Cyp11b-2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11B2, mitochondrial precursor (EC 1.14.15.4) (EC 1.14.15.5) (CYPXIB2) (P450C11) (Steroid 11-beta-hydroxylase) (Aldosterone synthase). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.004954 (rank : 25) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.004683 (rank : 26) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
GPV_MOUSE
|
||||||
| NC score | 0.003789 (rank : 27) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
EGR1_HUMAN
|
||||||
| NC score | 0.001752 (rank : 28) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||