Please be patient as the page loads
|
OSBP1_HUMAN
|
||||||
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OSBP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.974829 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBP2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.972015 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | 7.19216e-82 (rank : 4) | NC score | 0.623133 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL1_MOUSE
|
||||||
| θ value | 1.2268e-81 (rank : 5) | NC score | 0.637165 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL3_MOUSE
|
||||||
| θ value | 3.23591e-74 (rank : 6) | NC score | 0.889397 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL3_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 7) | NC score | 0.887020 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL2_MOUSE
|
||||||
| θ value | 1.15091e-71 (rank : 8) | NC score | 0.893362 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_HUMAN
|
||||||
| θ value | 6.31527e-70 (rank : 9) | NC score | 0.893286 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL6_MOUSE
|
||||||
| θ value | 1.31681e-67 (rank : 10) | NC score | 0.876192 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL7_HUMAN
|
||||||
| θ value | 1.45591e-66 (rank : 11) | NC score | 0.891433 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBL6_HUMAN
|
||||||
| θ value | 5.53243e-66 (rank : 12) | NC score | 0.878628 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSB11_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 13) | NC score | 0.693366 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB11_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 14) | NC score | 0.683649 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 15) | NC score | 0.676234 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PKHA3_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 16) | NC score | 0.580581 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PKHA3_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 17) | NC score | 0.564165 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |
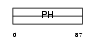
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 18) | NC score | 0.688193 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL9_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 19) | NC score | 0.716550 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 20) | NC score | 0.684812 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
C43BP_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 21) | NC score | 0.497453 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 22) | NC score | 0.500579 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
OSB10_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 23) | NC score | 0.692323 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.153967 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 25) | NC score | 0.200995 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 26) | NC score | 0.193034 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 27) | NC score | 0.121536 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 28) | NC score | 0.121220 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 29) | NC score | 0.099894 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 30) | NC score | 0.150783 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 31) | NC score | 0.158091 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 32) | NC score | 0.152365 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 33) | NC score | 0.156448 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 34) | NC score | 0.156638 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 35) | NC score | 0.109431 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 36) | NC score | 0.130087 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 37) | NC score | 0.142490 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 38) | NC score | 0.145550 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 39) | NC score | 0.129791 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 40) | NC score | 0.097652 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.053423 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.039519 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.151531 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 44) | NC score | 0.128160 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 45) | NC score | 0.053296 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 46) | NC score | 0.039730 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.154409 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
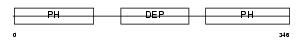
|
|||||
| Description | Pleckstrin-2. | |||||
|
URP2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.074442 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
AKT2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.011590 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31751, Q68GC0 | Gene names | AKT2 | |||
|
Domain Architecture |
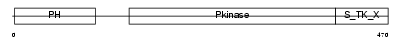
|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.125914 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.125908 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.121308 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
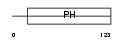
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.048396 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
URP2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 54) | NC score | 0.085491 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
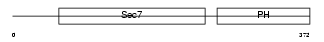
CYH1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.125220 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
DOCK9_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.049339 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
K1C10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.016727 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
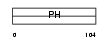
PKHB1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 58) | NC score | 0.119610 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.038540 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
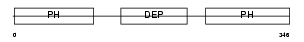
PLEK2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 60) | NC score | 0.153921 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
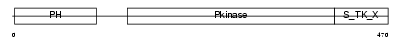
AKT1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.012538 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31750, Q62274 | Gene names | Akt1, Akt, Rac | |||
|
Domain Architecture |
|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (AKT1 kinase) (Protein kinase B) (PKB) (C-AKT) (Thymoma viral proto- oncogene). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.039080 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.163984 (rank : 63) | NC score | 0.024815 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.105950 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.103829 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.019497 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
AKT1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.012425 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P31749, Q9BWB6 | Gene names | AKT1, PKB, RAC | |||
|
Domain Architecture |
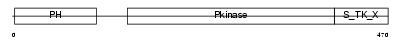
|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (Protein kinase B) (PKB) (C-AKT). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.123629 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.106793 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
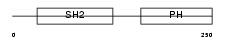
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 70) | NC score | 0.096932 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 71) | NC score | 0.038276 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.365318 (rank : 72) | NC score | 0.028708 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DOCK9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 73) | NC score | 0.037739 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.119347 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.036845 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.034443 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.068608 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRNT1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.042779 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q11, Q8ND57, Q9BS97, Q9Y362 | Gene names | TRNT1 | |||
|
Domain Architecture |
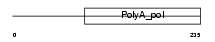
|
|||||
| Description | tRNA-nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.25) (Mitochondrial tRNA nucleotidyl transferase, CCA-adding) (mt tRNA adenylyltransferase) (mt tRNA CCA-pyrophosphorylase) (mt tRNA CCA- diphosphorylase) (mt CCA-adding enzyme). | |||||
|
TRNT1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.042824 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1J6, Q3TKW1, Q3UX99, Q920N6, Q9CSX0 | Gene names | Trnt1 | |||
|
Domain Architecture |
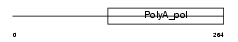
|
|||||
| Description | tRNA-nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.25) (mitochondrial tRNA nucleotidyl transferase, CCA-adding) (mt tRNA adenylyltransferase) (mt tRNA CCA-pyrophosphorylase) (mt tRNA CCA- diphosphorylase) (mt CCA-adding enzyme). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 80) | NC score | 0.028099 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
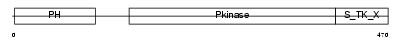
AKT2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.010961 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60823 | Gene names | Akt2 | |||
|
Domain Architecture |
|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.023694 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.039479 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.092972 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.018630 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.071134 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.055921 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.056527 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.054093 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.094406 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.038127 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.033606 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 93) | NC score | 0.017681 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 94) | NC score | 0.017544 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 95) | NC score | 0.033491 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 96) | NC score | 0.036407 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.053369 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CJ078_HUMAN
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.029898 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.027025 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
BAP29_HUMAN
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.031969 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||
|
CADH2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.001762 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 102) | NC score | 0.015955 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.035736 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.127673 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.120634 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.051581 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 107) | NC score | 0.031236 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.013252 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PROX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.032555 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
T3JAM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 110) | NC score | 0.039325 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.094340 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
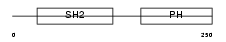
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
GAS8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 112) | NC score | 0.031971 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 113) | NC score | 0.038413 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 114) | NC score | 0.036343 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 115) | NC score | 0.038054 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PKHC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 116) | NC score | 0.041412 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
STX5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 117) | NC score | 0.023262 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1E0, Q3TM79, Q99J28, Q9JKP2 | Gene names | Stx5, Stx5a | |||
|
Domain Architecture |
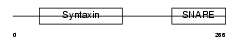
|
|||||
| Description | Syntaxin-5. | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.028628 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.005513 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.005811 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 121) | NC score | 0.050680 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 122) | NC score | 0.033757 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 123) | NC score | 0.042932 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.031995 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.008107 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.034766 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.054756 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.049041 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.028977 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.087559 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.020675 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PKHC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.040294 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.046079 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.004838 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
APOA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.019256 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02647, Q6LDN9, Q6Q785 | Gene names | APOA1 | |||
|
Domain Architecture |
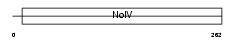
|
|||||
| Description | Apolipoprotein A-I precursor (Apo-AI) (ApoA-I) [Contains: Apolipoprotein A-I(1-242)]. | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.027679 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.103786 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.021943 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.024212 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.004976 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.017861 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.012882 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.023434 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.013793 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
TFE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.006524 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.028368 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.005320 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.005398 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.031153 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.021908 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.040036 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.029639 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.030687 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.015876 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LRP16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.013068 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |
|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.013294 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |
|
|||||
| Description | Protein LRP16. | |||||
|
LRRC6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.007008 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88978, Q9CUG2 | Gene names | Lrrc6, Lrtp, Mc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 6 (Leucine-rich testis-specific protein) (Testis-specific leucine-rich repeat protein). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.035210 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.118843 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.129857 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.005732 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.028568 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
T2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.015507 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
TEC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.000801 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.001343 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.072092 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.026926 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
EXOC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.016331 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R3S6 | Gene names | Exoc1, Sec3, Sec3l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.041321 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FRAT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.006980 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
K1C9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.014903 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.020977 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.036135 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.038756 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.006016 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.043808 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PKHB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.083382 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.002472 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.021630 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
ZN180_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | -0.005752 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJW8, Q9P1U2 | Gene names | ZNF180 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 180 (HHZ168). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.052774 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052661 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.069594 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.054176 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.090570 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.078016 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.090906 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.072927 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.069163 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.069166 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.058470 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.056865 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.127318 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PKHB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.086730 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.129187 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
PSD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.051350 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.061581 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.974829 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBP2_MOUSE
|
||||||
| NC score | 0.972015 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBL2_MOUSE
|
||||||
| NC score | 0.893362 (rank : 4) | θ value | 1.15091e-71 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_HUMAN
|
||||||
| NC score | 0.893286 (rank : 5) | θ value | 6.31527e-70 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL7_HUMAN
|
||||||
| NC score | 0.891433 (rank : 6) | θ value | 1.45591e-66 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBL3_MOUSE
|
||||||
| NC score | 0.889397 (rank : 7) | θ value | 3.23591e-74 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL3_HUMAN
|
||||||
| NC score | 0.887020 (rank : 8) | θ value | 4.67263e-73 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL6_HUMAN
|
||||||
| NC score | 0.878628 (rank : 9) | θ value | 5.53243e-66 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL6_MOUSE
|
||||||
| NC score | 0.876192 (rank : 10) | θ value | 1.31681e-67 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL9_HUMAN
|
||||||
| NC score | 0.716550 (rank : 11) | θ value | 3.28887e-18 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSB11_MOUSE
|
||||||
| NC score | 0.693366 (rank : 12) | θ value | 2.43343e-21 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB10_HUMAN
|
||||||
| NC score | 0.692323 (rank : 13) | θ value | 4.16044e-13 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.688193 (rank : 14) | θ value | 2.5182e-18 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.684812 (rank : 15) | θ value | 1.63225e-17 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
OSB11_HUMAN
|
||||||
| NC score | 0.683649 (rank : 16) | θ value | 4.15078e-21 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.676234 (rank : 17) | θ value | 5.42112e-21 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
OSBL1_MOUSE
|
||||||
| NC score | 0.637165 (rank : 18) | θ value | 1.2268e-81 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.623133 (rank : 19) | θ value | 7.19216e-82 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
PKHA3_MOUSE
|
||||||
| NC score | 0.580581 (rank : 20) | θ value | 2.27762e-19 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PKHA3_HUMAN
|
||||||
| NC score | 0.564165 (rank : 21) | θ value | 2.97466e-19 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
C43BP_HUMAN
|
||||||
| NC score | 0.500579 (rank : 22) | θ value | 7.58209e-15 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_MOUSE
|
||||||
| NC score | 0.497453 (rank : 23) | θ value | 4.44505e-15 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.200995 (rank : 24) | θ value | 3.19293e-05 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.193034 (rank : 25) | θ value | 7.1131e-05 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.158091 (rank : 26) | θ value | 0.00102713 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.156638 (rank : 27) | θ value | 0.00228821 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.156448 (rank : 28) | θ value | 0.00175202 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.154409 (rank : 29) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.153967 (rank : 30) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.153921 (rank : 31) | θ value | 0.125558 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.152365 (rank : 32) | θ value | 0.00134147 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.151531 (rank : 33) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.150783 (rank : 34) | θ value | 0.000786445 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.145550 (rank : 35) | θ value | 0.0113563 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.142490 (rank : 36) | θ value | 0.0113563 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.130087 (rank : 37) | θ value | 0.00665767 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.129857 (rank : 38) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.129791 (rank : 39) | θ value | 0.0148317 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.129187 (rank : 40) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.128160 (rank : 41) | θ value | 0.0563607 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.127673 (rank : 42) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.127318 (rank : 43) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.125914 (rank : 44) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.125908 (rank : 45) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.125220 (rank : 46) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.123629 (rank : 47) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.121536 (rank : 48) | θ value | 9.29e-05 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.121308 (rank : 49) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.121220 (rank : 50) | θ value | 0.000121331 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.120634 (rank : 51) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHB1_MOUSE
|
||||||
| NC score | 0.119610 (rank : 52) | θ value | 0.0961366 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.119347 (rank : 53) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.118843 (rank : 54) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.109431 (rank : 55) | θ value | 0.00390308 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.106793 (rank : 56) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.105950 (rank : 57) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.103829 (rank : 58) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.103786 (rank : 59) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.099894 (rank : 60) | θ value | 0.000121331 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.097652 (rank : 61) | θ value | 0.0252991 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.096932 (rank : 62) | θ value | 0.279714 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.094406 (rank : 63) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.094340 (rank : 64) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.092972 (rank : 65) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.090906 (rank : 66) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.090570 (rank : 67) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.087559 (rank : 68) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PKHB2_MOUSE
|
||||||
| NC score | 0.086730 (rank : 69) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
URP2_MOUSE
|
||||||
| NC score | 0.085491 (rank : 70) | θ value | 0.0736092 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
PKHB2_HUMAN
|
||||||
| NC score | 0.083382 (rank : 71) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.078016 (rank : 72) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
URP2_HUMAN
|
||||||
| NC score | 0.074442 (rank : 73) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.072927 (rank : 74) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.072092 (rank : 75) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.071134 (rank : 76) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.069594 (rank : 77) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.069166 (rank : 78) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.069163 (rank : 79) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.068608 (rank : 80) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.061581 (rank : 81) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.058470 (rank : 82) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.056865 (rank : 83) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.056527 (rank : 84) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.055921 (rank : 85) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.054756 (rank : 86) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.054176 (rank : 87) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.054093 (rank : 88) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.053423 (rank : 89) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.053369 (rank : 90) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.053296 (rank : 91) | θ value | 0.0563607 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.052774 (rank : 92) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.052661 (rank : 93) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.051581 (rank : 94) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
PSD3_HUMAN
|
||||||
| NC score | 0.051350 (rank : 95) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.050680 (rank : 96) | θ value | 3.0926 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
DOCK9_HUMAN
|
||||||
| NC score | 0.049339 (rank : 97) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.049041 (rank : 98) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.048396 (rank : 99) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.046079 (rank : 100) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.043808 (rank : 101) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.042932 (rank : 102) | θ value | 3.0926 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
TRNT1_MOUSE
|
||||||
| NC score | 0.042824 (rank : 103) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1J6, Q3TKW1, Q3UX99, Q920N6, Q9CSX0 | Gene names | Trnt1 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA-nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.25) (mitochondrial tRNA nucleotidyl transferase, CCA-adding) (mt tRNA adenylyltransferase) (mt tRNA CCA-pyrophosphorylase) (mt tRNA CCA- diphosphorylase) (mt CCA-adding enzyme). | |||||
|
TRNT1_HUMAN
|
||||||
| NC score | 0.042779 (rank : 104) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q11, Q8ND57, Q9BS97, Q9Y362 | Gene names | TRNT1 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA-nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.25) (Mitochondrial tRNA nucleotidyl transferase, CCA-adding) (mt tRNA adenylyltransferase) (mt tRNA CCA-pyrophosphorylase) (mt tRNA CCA- diphosphorylase) (mt CCA-adding enzyme). | |||||
|
PKHC1_HUMAN
|
||||||
| NC score | 0.041412 (rank : 105) | θ value | 2.36792 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.041321 (rank : 106) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
PKHC1_MOUSE
|
||||||
| NC score | 0.040294 (rank : 107) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.040036 (rank : 108) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.039730 (rank : 109) | θ value | 0.0563607 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.039519 (rank : 110) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.039479 (rank : 111) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
T3JAM_HUMAN
|
||||||
| NC score | 0.039325 (rank : 112) | θ value | 1.81305 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.039080 (rank : 113) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.038756 (rank : 114) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.038540 (rank : 115) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.038413 (rank : 116) | θ value | 2.36792 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.038276 (rank : 117) | θ value | 0.279714 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.038127 (rank : 118) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.038054 (rank : 119) | θ value | 2.36792 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
DOCK9_MOUSE
|
||||||
| NC score | 0.037739 (rank : 120) | θ value | 0.365318 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.036845 (rank : 121) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.036407 (rank : 122) | θ value | 0.813845 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.036343 (rank : 123) | θ value | 2.36792 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.036135 (rank : 124) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.035736 (rank : 125) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.035210 (rank : 126) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.034766 (rank : 127) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.034443 (rank : 128) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.033757 (rank : 129) | θ value | 3.0926 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.033606 (rank : 130) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.033491 (rank : 131) | θ value | 0.813845 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
PROX1_HUMAN
|
||||||
| NC score | 0.032555 (rank : 132) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.031995 (rank : 133) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.031971 (rank : 134) | θ value | 2.36792 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
BAP29_HUMAN
|
||||||
| NC score | 0.031969 (rank : 135) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.031236 (rank : 136) | θ value | 1.81305 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.031153 (rank : 137) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.030687 (rank : 138) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
CJ078_HUMAN
|
||||||
| NC score | 0.029898 (rank : 139) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.029639 (rank : 140) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.028977 (rank : 141) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.028708 (rank : 142) | θ value | 0.365318 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.028628 (rank : 143) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.028568 (rank : 144) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.028368 (rank : 145) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.028099 (rank : 146) | θ value | 0.365318 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.027679 (rank : 147) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.027025 (rank : 148) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.026926 (rank : 149) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.024815 (rank : 150) | θ value | 0.163984 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.024212 (rank : 151) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.023694 (rank : 152) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.023434 (rank : 153) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
STX5_MOUSE
|
||||||
| NC score | 0.023262 (rank : 154) | θ value | 2.36792 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1E0, Q3TM79, Q99J28, Q9JKP2 | Gene names | Stx5, Stx5a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-5. | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.021943 (rank : 155) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.021908 (rank : 156) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.021630 (rank : 157) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.020977 (rank : 158) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.020675 (rank : 159) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.019497 (rank : 160) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
APOA1_HUMAN
|
||||||
| NC score | 0.019256 (rank : 161) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02647, Q6LDN9, Q6Q785 | Gene names | APOA1 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-I precursor (Apo-AI) (ApoA-I) [Contains: Apolipoprotein A-I(1-242)]. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.018630 (rank : 162) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.017861 (rank : 163) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.017681 (rank : 164) | θ value | 0.813845 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.017544 (rank : 165) | θ value | 0.813845 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
K1C10_MOUSE
|
||||||
| NC score | 0.016727 (rank : 166) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
EXOC1_MOUSE
|
||||||
| NC score | 0.016331 (rank : 167) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R3S6 | Gene names | Exoc1, Sec3, Sec3l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.015955 (rank : 168) | θ value | 1.38821 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.015876 (rank : 169) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.015507 (rank : 170) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
K1C9_HUMAN
|
||||||
| NC score | 0.014903 (rank : 171) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.013793 (rank : 172) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
LRP16_MOUSE
|
||||||
| NC score | 0.013294 (rank : 173) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.013252 (rank : 174) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
LRP16_HUMAN
|
||||||
| NC score | 0.013068 (rank : 175) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.012882 (rank : 176) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
AKT1_MOUSE
|
||||||
| NC score | 0.012538 (rank : 177) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31750, Q62274 | Gene names | Akt1, Akt, Rac | |||
|
Domain Architecture |

|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (AKT1 kinase) (Protein kinase B) (PKB) (C-AKT) (Thymoma viral proto- oncogene). | |||||
|
AKT1_HUMAN
|
||||||
| NC score | 0.012425 (rank : 178) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P31749, Q9BWB6 | Gene names | AKT1, PKB, RAC | |||
|
Domain Architecture |

|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (Protein kinase B) (PKB) (C-AKT). | |||||
|
AKT2_HUMAN
|
||||||
| NC score | 0.011590 (rank : 179) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31751, Q68GC0 | Gene names | AKT2 | |||
|
Domain Architecture |

|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||
|
AKT2_MOUSE
|
||||||
| NC score | 0.010961 (rank : 180) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60823 | Gene names | Akt2 | |||
|
Domain Architecture |

|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.008107 (rank : 181) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
LRRC6_MOUSE
|
||||||
| NC score | 0.007008 (rank : 182) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88978, Q9CUG2 | Gene names | Lrrc6, Lrtp, Mc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 6 (Leucine-rich testis-specific protein) (Testis-specific leucine-rich repeat protein). | |||||
|
FRAT1_HUMAN
|
||||||
| NC score | 0.006980 (rank : 183) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.006524 (rank : 184) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.006016 (rank : 185) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.005811 (rank : 186) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.005732 (rank : 187) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.005513 (rank : 188) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.005398 (rank : 189) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.005320 (rank : 190) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.004976 (rank : 191) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.004838 (rank : 192) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.002472 (rank : 193) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CADH2_MOUSE
|
||||||
| NC score | 0.001762 (rank : 194) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.001343 (rank : 195) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
TEC_MOUSE
|
||||||
| NC score | 0.000801 (rank : 196) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
ZN180_HUMAN
|
||||||
| NC score | -0.005752 (rank : 197) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJW8, Q9P1U2 | Gene names | ZNF180 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 180 (HHZ168). | |||||