Please be patient as the page loads
|
MCCA_HUMAN
|
||||||
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCCA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998357 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 4.44261e-132 (rank : 3) | NC score | 0.966541 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 1.33764e-128 (rank : 4) | NC score | 0.965229 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 1.34698e-104 (rank : 5) | NC score | 0.929228 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| θ value | 2.2976e-104 (rank : 6) | NC score | 0.929332 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 6.55044e-59 (rank : 7) | NC score | 0.821243 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| θ value | 1.51013e-55 (rank : 8) | NC score | 0.785866 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 9) | NC score | 0.414289 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 10) | NC score | 0.407599 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 11) | NC score | 0.391790 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.124514 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
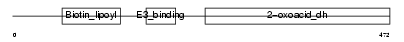
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.118943 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
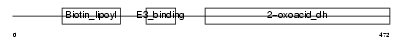
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.090070 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODPX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.087384 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
PUR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.060409 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.000789 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.004191 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
SRY_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.006357 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.007578 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.007548 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.012165 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.011464 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
ZN406_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | -0.000971 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.005950 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
PUR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.043002 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
HMGCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.054558 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |
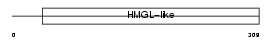
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
HMGCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.062424 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |
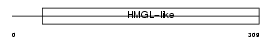
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PCCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.086782 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.089358 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.998357 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.966541 (rank : 3) | θ value | 4.44261e-132 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.965229 (rank : 4) | θ value | 1.33764e-128 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.929332 (rank : 5) | θ value | 2.2976e-104 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.929228 (rank : 6) | θ value | 1.34698e-104 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.821243 (rank : 7) | θ value | 6.55044e-59 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.785866 (rank : 8) | θ value | 1.51013e-55 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.414289 (rank : 9) | θ value | 1.47974e-10 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.407599 (rank : 10) | θ value | 1.25267e-09 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.391790 (rank : 11) | θ value | 1.99992e-07 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.124514 (rank : 12) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.118943 (rank : 13) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.090070 (rank : 14) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
PCCB_MOUSE
|
||||||
| NC score | 0.089358 (rank : 15) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.087384 (rank : 16) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
PCCB_HUMAN
|
||||||
| NC score | 0.086782 (rank : 17) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
HMGCL_MOUSE
|
||||||
| NC score | 0.062424 (rank : 18) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PUR2_HUMAN
|
||||||
| NC score | 0.060409 (rank : 19) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
HMGCL_HUMAN
|
||||||
| NC score | 0.054558 (rank : 20) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PUR2_MOUSE
|
||||||
| NC score | 0.043002 (rank : 21) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.012165 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.011464 (rank : 23) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.007578 (rank : 24) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.007548 (rank : 25) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.006357 (rank : 26) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.005950 (rank : 27) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.004191 (rank : 28) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.000789 (rank : 29) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN406_HUMAN
|
||||||
| NC score | -0.000971 (rank : 30) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||