Please be patient as the page loads
|
PYC_HUMAN
|
||||||
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PYC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999139 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 1.34698e-104 (rank : 3) | NC score | 0.930919 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 2.2976e-104 (rank : 4) | NC score | 0.929332 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 6.68496e-104 (rank : 5) | NC score | 0.931006 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 2.46047e-98 (rank : 6) | NC score | 0.926608 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 6.13105e-57 (rank : 7) | NC score | 0.795358 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| θ value | 2.1121e-49 (rank : 8) | NC score | 0.759553 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 9) | NC score | 0.403585 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 10) | NC score | 0.404860 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 11) | NC score | 0.397388 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
HMGCL_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 12) | NC score | 0.160698 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |
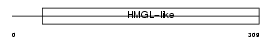
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
HMGCL_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.148015 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |
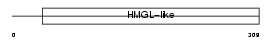
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.019226 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
NEK11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.000221 (rank : 27) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0Q4, Q8BLS6, Q8BW62 | Gene names | Nek11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
HEM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.028766 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13716, Q16870, Q16871, Q9BVQ9 | Gene names | ALAD | |||
|
Domain Architecture |
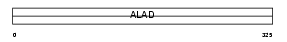
|
|||||
| Description | Delta-aminolevulinic acid dehydratase (EC 4.2.1.24) (Porphobilinogen synthase) (ALADH). | |||||
|
CPNE7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.006948 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.026516 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.026570 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.009708 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
LNK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.014683 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |
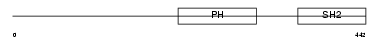
|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
PUR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.043671 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
CARD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.003437 (rank : 26) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H257, Q5SXM6, Q9H854 | Gene names | CARD9 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 9 (hCARD9). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.010890 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.005759 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PCCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.080748 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.083158 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.999139 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.931006 (rank : 3) | θ value | 6.68496e-104 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.930919 (rank : 4) | θ value | 1.34698e-104 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.929332 (rank : 5) | θ value | 2.2976e-104 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.926608 (rank : 6) | θ value | 2.46047e-98 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.795358 (rank : 7) | θ value | 6.13105e-57 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.759553 (rank : 8) | θ value | 2.1121e-49 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.404860 (rank : 9) | θ value | 8.11959e-09 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.403585 (rank : 10) | θ value | 4.59992e-12 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.397388 (rank : 11) | θ value | 4.0297e-08 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
HMGCL_MOUSE
|
||||||
| NC score | 0.160698 (rank : 12) | θ value | 0.00175202 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
HMGCL_HUMAN
|
||||||
| NC score | 0.148015 (rank : 13) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PCCB_MOUSE
|
||||||
| NC score | 0.083158 (rank : 14) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_HUMAN
|
||||||
| NC score | 0.080748 (rank : 15) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PUR2_HUMAN
|
||||||
| NC score | 0.043671 (rank : 16) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
HEM2_HUMAN
|
||||||
| NC score | 0.028766 (rank : 17) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13716, Q16870, Q16871, Q9BVQ9 | Gene names | ALAD | |||
|
Domain Architecture |

|
|||||
| Description | Delta-aminolevulinic acid dehydratase (EC 4.2.1.24) (Porphobilinogen synthase) (ALADH). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.026570 (rank : 18) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.026516 (rank : 19) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.019226 (rank : 20) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
LNK_HUMAN
|
||||||
| NC score | 0.014683 (rank : 21) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.010890 (rank : 22) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.009708 (rank : 23) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CPNE7_HUMAN
|
||||||
| NC score | 0.006948 (rank : 24) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.005759 (rank : 25) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
CARD9_HUMAN
|
||||||
| NC score | 0.003437 (rank : 26) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H257, Q5SXM6, Q9H854 | Gene names | CARD9 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 9 (hCARD9). | |||||
|
NEK11_MOUSE
|
||||||
| NC score | 0.000221 (rank : 27) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0Q4, Q8BLS6, Q8BW62 | Gene names | Nek11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||