Please be patient as the page loads
|
SACS_MOUSE
|
||||||
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SACS_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994673 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 3) | NC score | 0.190831 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
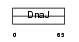
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 4) | NC score | 0.191589 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.186571 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
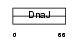
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 6) | NC score | 0.184834 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
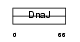
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.188982 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
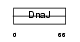
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.184087 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB7_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.185144 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.032702 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DNJB3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.184321 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 12) | NC score | 0.194644 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJC5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.168803 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.168776 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.157721 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.124893 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.123574 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.137978 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
DNJ5B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.149985 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.127484 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
O51A4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.004099 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGJ6 | Gene names | OR51A4 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 51A4. | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.126383 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.125366 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
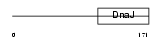
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
ENPL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.044892 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.022397 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
HEAT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.070758 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.026387 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.042625 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.033929 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
COMDA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.076372 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZY2 | Gene names | Commd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 10 (Down-regulated in W/WV mouse stomach 2) (mDRWMS2). | |||||
|
ENPL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.043053 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.020951 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.025467 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
MOR2B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.037560 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.120853 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.120707 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.016605 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
OTOAN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.043538 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.121374 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.124796 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.116104 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
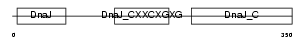
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.116234 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
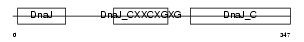
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
PERT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.024503 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
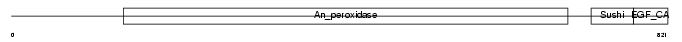
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
POLK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.035840 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.046885 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.013901 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CUL4A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.025249 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.149886 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.043587 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
PDZD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.049628 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
PI3R5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.023502 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WYR1, Q5G936, Q5G938, Q5G939, Q8IZ23, Q9Y2Y2 | Gene names | PIK3R5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit) (Protein FOAP-2). | |||||
|
PP2CD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.021529 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
TLR13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.010786 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
A2MG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.015186 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01023, Q13677, Q59F47, Q5QTS0, Q68DN2, Q6PIY3, Q6PN97 | Gene names | A2M | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M). | |||||
|
CN094_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.037825 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6D7, Q86T15, Q86T16, Q86U43, Q9NX59 | Gene names | C14orf94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf94. | |||||
|
COMDA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.060128 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6G5, Q9P077 | Gene names | COMMD10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 10. | |||||
|
SYMPK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.041878 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
TTLL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.028281 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
TTLL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.028329 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
CCD13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.017870 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.018756 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
GEMI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.039409 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQQ4, Q9DAD7 | Gene names | Sip1, Gemin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Survival of motor neuron protein-interacting protein 1 (SMN- interacting protein 1) (Component of gems 2) (Gemin-2). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.012316 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.017629 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.011015 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.013920 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PYC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.026570 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.026503 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.021498 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SRXN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.054329 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D975, Q3TWN6, Q91VN5 | Gene names | Srxn1, Npn3, Srx | |||
|
Domain Architecture |
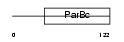
|
|||||
| Description | Sulfiredoxin-1 (EC 1.8.98.2) (Neoplastic progression protein 3). | |||||
|
STAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.031382 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.011631 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
DJC16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.103677 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.115030 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
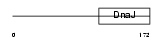
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.012814 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.017919 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
O51A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.001945 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGJ7 | Gene names | OR51A2 | |||
|
Domain Architecture |
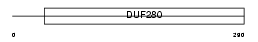
|
|||||
| Description | Olfactory receptor 51A2. | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.020307 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
TAOK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.004501 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.004406 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.010724 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ABLM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.004765 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
AVEN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.031268 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9K3 | Gene names | Aven | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.006763 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
HEM3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.031170 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22907 | Gene names | Hmbs, Uros1 | |||
|
Domain Architecture |
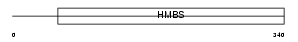
|
|||||
| Description | Porphobilinogen deaminase (EC 2.5.1.61) (Hydroxymethylbilane synthase) (HMBS) (Pre-uroporphyrinogen synthase) (PBG-D). | |||||
|
K1219_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018820 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
MPP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.010020 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.013954 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.010944 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.020281 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.006334 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.011026 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
CP070_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.012942 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSU1 | Gene names | C16orf70 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0183 protein C16orf70. | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.012666 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.089194 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.087500 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DJC18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.105467 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
MELPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.016734 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.000598 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CU055_HUMAN
|
||||||
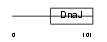
| θ value | θ > 10 (rank : 100) | NC score | 0.050529 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
CU055_MOUSE
|
||||||
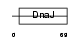
| θ value | θ > 10 (rank : 101) | NC score | 0.056645 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.090289 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.089351 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.081542 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC12_MOUSE
|
||||||
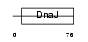
| θ value | θ > 10 (rank : 105) | NC score | 0.081874 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.091094 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.091918 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJA3_HUMAN
|
||||||
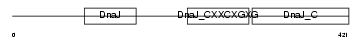
| θ value | θ > 10 (rank : 108) | NC score | 0.098033 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.098900 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 31 | |
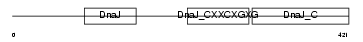
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.097218 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.097446 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.097842 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.097901 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.100235 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
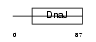
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.100627 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
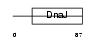
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.098425 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.098563 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.097928 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.097344 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.085102 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.086338 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.073805 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.073678 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.088984 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 31 | |
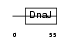
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.086162 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
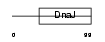
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.071258 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.070293 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.071892 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.071393 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.090338 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.090614 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.059679 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.059376 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
WBS18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.091980 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
WBS18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.096768 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.079338 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.072946 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.994673 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.194644 (rank : 3) | θ value | 0.0113563 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.191589 (rank : 4) | θ value | 0.00134147 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.190831 (rank : 5) | θ value | 0.00102713 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.188982 (rank : 6) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.186571 (rank : 7) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.185144 (rank : 8) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.184834 (rank : 9) | θ value | 0.00390308 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.184321 (rank : 10) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.184087 (rank : 11) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.168803 (rank : 12) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.168776 (rank : 13) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.157721 (rank : 14) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.149985 (rank : 15) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.149886 (rank : 16) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.137978 (rank : 17) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.127484 (rank : 18) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.126383 (rank : 19) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.125366 (rank : 20) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.124893 (rank : 21) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.124796 (rank : 22) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.123574 (rank : 23) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.121374 (rank : 24) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.120853 (rank : 25) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.120707 (rank : 26) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.116234 (rank : 27) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.116104 (rank : 28) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.115030 (rank : 29) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.105467 (rank : 30) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.103677 (rank : 31) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.100627 (rank : 32) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.100235 (rank : 33) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.098900 (rank : 34) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.098563 (rank : 35) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.098425 (rank : 36) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.098033 (rank : 37) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.097928 (rank : 38) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.097901 (rank : 39) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.097842 (rank : 40) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.097446 (rank : 41) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.097344 (rank : 42) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.097218 (rank : 43) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.096768 (rank : 44) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.091980 (rank : 45) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.091918 (rank : 46) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.091094 (rank : 47) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.090614 (rank : 48) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.090338 (rank : 49) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.090289 (rank : 50) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.089351 (rank : 51) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.089194 (rank : 52) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.088984 (rank : 53) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.087500 (rank : 54) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.086338 (rank : 55) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.086162 (rank : 56) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.085102 (rank : 57) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.081874 (rank : 58) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.081542 (rank : 59) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.079338 (rank : 60) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
COMDA_MOUSE
|
||||||
| NC score | 0.076372 (rank : 61) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZY2 | Gene names | Commd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 10 (Down-regulated in W/WV mouse stomach 2) (mDRWMS2). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.073805 (rank : 62) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.073678 (rank : 63) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.072946 (rank : 64) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.071892 (rank : 65) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.071393 (rank : 66) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.071258 (rank : 67) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
HEAT1_HUMAN
|
||||||
| NC score | 0.070758 (rank : 68) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.070293 (rank : 69) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
COMDA_HUMAN
|
||||||
| NC score | 0.060128 (rank : 70) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6G5, Q9P077 | Gene names | COMMD10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 10. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.059679 (rank : 71) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.059376 (rank : 72) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.056645 (rank : 73) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
SRXN1_MOUSE
|
||||||
| NC score | 0.054329 (rank : 74) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D975, Q3TWN6, Q91VN5 | Gene names | Srxn1, Npn3, Srx | |||
|
Domain Architecture |

|
|||||
| Description | Sulfiredoxin-1 (EC 1.8.98.2) (Neoplastic progression protein 3). | |||||
|
CU055_HUMAN
|
||||||
| NC score | 0.050529 (rank : 75) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
PDZD6_HUMAN
|
||||||
| NC score | 0.049628 (rank : 76) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.046885 (rank : 77) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
ENPL_HUMAN
|
||||||
| NC score | 0.044892 (rank : 78) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.043587 (rank : 79) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
OTOAN_MOUSE
|
||||||
| NC score | 0.043538 (rank : 80) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
ENPL_MOUSE
|
||||||
| NC score | 0.043053 (rank : 81) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.042625 (rank : 82) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.041878 (rank : 83) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
GEMI2_MOUSE
|
||||||
| NC score | 0.039409 (rank : 84) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQQ4, Q9DAD7 | Gene names | Sip1, Gemin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Survival of motor neuron protein-interacting protein 1 (SMN- interacting protein 1) (Component of gems 2) (Gemin-2). | |||||
|
CN094_HUMAN
|
||||||
| NC score | 0.037825 (rank : 85) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6D7, Q86T15, Q86T16, Q86U43, Q9NX59 | Gene names | C14orf94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf94. | |||||
|
MOR2B_MOUSE
|
||||||
| NC score | 0.037560 (rank : 86) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
POLK_MOUSE
|
||||||
| NC score | 0.035840 (rank : 87) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.033929 (rank : 88) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.032702 (rank : 89) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.031382 (rank : 90) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
AVEN_MOUSE
|
||||||
| NC score | 0.031268 (rank : 91) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9K3 | Gene names | Aven | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
HEM3_MOUSE
|
||||||
| NC score | 0.031170 (rank : 92) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22907 | Gene names | Hmbs, Uros1 | |||
|
Domain Architecture |

|
|||||
| Description | Porphobilinogen deaminase (EC 2.5.1.61) (Hydroxymethylbilane synthase) (HMBS) (Pre-uroporphyrinogen synthase) (PBG-D). | |||||
|
TTLL1_MOUSE
|
||||||
| NC score | 0.028329 (rank : 93) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
TTLL1_HUMAN
|
||||||
| NC score | 0.028281 (rank : 94) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.026570 (rank : 95) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.026503 (rank : 96) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.026387 (rank : 97) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.025467 (rank : 98) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.025249 (rank : 99) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.024503 (rank : 100) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
PI3R5_HUMAN
|
||||||
| NC score | 0.023502 (rank : 101) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WYR1, Q5G936, Q5G938, Q5G939, Q8IZ23, Q9Y2Y2 | Gene names | PIK3R5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit) (Protein FOAP-2). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.022397 (rank : 102) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.021529 (rank : 103) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.021498 (rank : 104) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.020951 (rank : 105) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.020307 (rank : 106) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.020281 (rank : 107) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
K1219_MOUSE
|
||||||
| NC score | 0.018820 (rank : 108) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.018756 (rank : 109) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.017919 (rank : 110) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.017870 (rank : 111) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.017629 (rank : 112) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.016734 (rank : 113) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.016605 (rank : 114) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
A2MG_HUMAN
|
||||||
| NC score | 0.015186 (rank : 115) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01023, Q13677, Q59F47, Q5QTS0, Q68DN2, Q6PIY3, Q6PN97 | Gene names | A2M | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.013954 (rank : 116) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.013920 (rank : 117) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.013901 (rank : 118) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CP070_HUMAN
|
||||||
| NC score | 0.012942 (rank : 119) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSU1 | Gene names | C16orf70 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0183 protein C16orf70. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.012814 (rank : 120) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.012666 (rank : 121) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.012316 (rank : 122) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.011631 (rank : 123) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.011026 (rank : 124) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.011015 (rank : 125) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.010944 (rank : 126) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
TLR13_MOUSE
|
||||||
| NC score | 0.010786 (rank : 127) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.010724 (rank : 128) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MPP4_MOUSE
|
||||||
| NC score | 0.010020 (rank : 129) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.006763 (rank : 130) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.006334 (rank : 131) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
ABLM2_HUMAN
|
||||||
| NC score | 0.004765 (rank : 132) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
TAOK1_HUMAN
|
||||||
| NC score | 0.004501 (rank : 133) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.004406 (rank : 134) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
O51A4_HUMAN
|
||||||
| NC score | 0.004099 (rank : 135) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGJ6 | Gene names | OR51A4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 51A4. | |||||
|
O51A2_HUMAN
|
||||||
| NC score | 0.001945 (rank : 136) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGJ7 | Gene names | OR51A2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 51A2. | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.000598 (rank : 137) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||