Please be patient as the page loads
|
DJC12_HUMAN
|
||||||
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DJC12_HUMAN
|
||||||
| θ value | 4.1775e-114 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC12_MOUSE
|
||||||
| θ value | 2.64101e-84 (rank : 2) | NC score | 0.969658 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 3) | NC score | 0.749584 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 4) | NC score | 0.769756 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 5) | NC score | 0.730759 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 6) | NC score | 0.730344 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 7) | NC score | 0.733809 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJC5_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 8) | NC score | 0.744773 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 9) | NC score | 0.744656 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
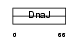
DNJB7_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.721639 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DJC16_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 11) | NC score | 0.691061 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 12) | NC score | 0.683615 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
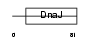
DNJ5B_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 13) | NC score | 0.741422 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 14) | NC score | 0.731632 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
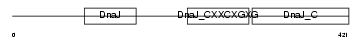
DNJA3_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 15) | NC score | 0.720220 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
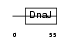
DNJC4_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 16) | NC score | 0.730701 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.739860 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
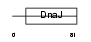
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 18) | NC score | 0.730703 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
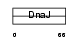
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DJC16_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 19) | NC score | 0.685714 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.714240 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
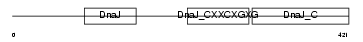
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.682577 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 22) | NC score | 0.682587 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 23) | NC score | 0.682473 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 24) | NC score | 0.682212 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 25) | NC score | 0.730161 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
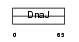
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 26) | NC score | 0.709583 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 27) | NC score | 0.677127 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 28) | NC score | 0.721463 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 29) | NC score | 0.675719 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 30) | NC score | 0.704010 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 31) | NC score | 0.704963 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB3_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 32) | NC score | 0.715783 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
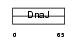
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 33) | NC score | 0.717229 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
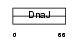
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DJC18_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 34) | NC score | 0.693307 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 35) | NC score | 0.548700 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 36) | NC score | 0.546618 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 37) | NC score | 0.665931 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 38) | NC score | 0.711053 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
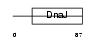
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 39) | NC score | 0.579030 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 40) | NC score | 0.577404 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 41) | NC score | 0.663885 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 42) | NC score | 0.657938 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 43) | NC score | 0.656789 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 44) | NC score | 0.644262 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 45) | NC score | 0.641162 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 46) | NC score | 0.705995 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
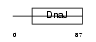
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 47) | NC score | 0.605703 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 48) | NC score | 0.602178 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 49) | NC score | 0.647566 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
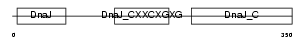
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 50) | NC score | 0.647736 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
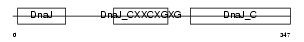
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 51) | NC score | 0.645027 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 52) | NC score | 0.683338 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 53) | NC score | 0.642142 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 54) | NC score | 0.604026 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 55) | NC score | 0.657553 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 56) | NC score | 0.694095 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 57) | NC score | 0.650113 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 58) | NC score | 0.057258 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
WBS18_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 59) | NC score | 0.671451 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 60) | NC score | 0.618654 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 61) | NC score | 0.619965 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 62) | NC score | 0.627167 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.012704 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 64) | NC score | 0.444553 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 65) | NC score | 0.448207 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
WBS18_HUMAN
|
||||||
| θ value | 0.125558 (rank : 66) | NC score | 0.667798 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 67) | NC score | 0.033149 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 68) | NC score | 0.029122 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.032622 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CU055_MOUSE
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.407394 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.063437 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 0.365318 (rank : 72) | NC score | 0.033134 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 73) | NC score | 0.014438 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.490673 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.489358 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.048143 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 77) | NC score | 0.046759 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.019177 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.038533 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.035379 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.042280 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.027266 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.030945 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.012233 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.023390 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.032553 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.010699 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.036619 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.028383 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.035101 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.026481 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.019688 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.029492 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.021720 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
ING4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.025414 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.028660 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
SPC25_MOUSE
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.037214 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.044597 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.015923 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.011041 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
K1718_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.012576 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MCM8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.012236 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
EXOSX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.019274 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
ING4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.022492 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.033648 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.009524 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.026165 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.010614 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ENPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.018515 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.019347 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.024052 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.021403 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.021017 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.029835 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TMCO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.031451 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM00, O75545, Q9BZS3, Q9BZU8 | Gene names | TMCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 1 (Xenogeneic cross-immune protein PCIA3). | |||||
|
TMCO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.031451 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921L3, Q3TS11 | Gene names | Tmco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 1. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.011462 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.014946 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.029590 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.004843 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.032000 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.037274 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.031090 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.025166 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.015969 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.011830 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.031162 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.031571 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.009804 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.014366 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
K1C13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.004873 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
KNG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.013249 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01042, P01043 | Gene names | KNG1, BDK, KNG | |||
|
Domain Architecture |
|
|||||
| Description | Kininogen-1 precursor (Alpha-2-thiol proteinase inhibitor) [Contains: Kininogen-1 heavy chain; Bradykinin (Kallidin I); Lysyl-bradykinin (Kallidin II); Kininogen-1 light chain; Low molecular weight growth- promoting factor]. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | -0.001117 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.018198 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.029017 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PDCL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.010568 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.001301 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.026943 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.000646 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.024058 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.321893 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
AUXI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.082631 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.072594 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
CU055_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.372618 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
DCJ15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.095588 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
DCJ15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.077018 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.337304 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
SACS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.080710 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.081542 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
TIM14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.100805 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.100323 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056602 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.303395 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.1775e-114 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.969658 (rank : 2) | θ value | 2.64101e-84 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.769756 (rank : 3) | θ value | 2.43908e-13 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.749584 (rank : 4) | θ value | 9.90251e-15 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.744773 (rank : 5) | θ value | 4.45536e-07 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.744656 (rank : 6) | θ value | 4.45536e-07 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.741422 (rank : 7) | θ value | 7.59969e-07 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.739860 (rank : 8) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.733809 (rank : 9) | θ value | 6.87365e-08 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.731632 (rank : 10) | θ value | 7.59969e-07 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.730759 (rank : 11) | θ value | 1.80886e-08 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.730703 (rank : 12) | θ value | 2.88788e-06 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.730701 (rank : 13) | θ value | 1.69304e-06 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.730344 (rank : 14) | θ value | 1.80886e-08 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.730161 (rank : 15) | θ value | 6.43352e-06 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.721639 (rank : 16) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.721463 (rank : 17) | θ value | 2.44474e-05 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.720220 (rank : 18) | θ value | 1.29631e-06 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.717229 (rank : 19) | θ value | 5.44631e-05 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.715783 (rank : 20) | θ value | 4.1701e-05 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.714240 (rank : 21) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.711053 (rank : 22) | θ value | 0.00035302 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.709583 (rank : 23) | θ value | 1.43324e-05 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.705995 (rank : 24) | θ value | 0.00134147 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.704963 (rank : 25) | θ value | 4.1701e-05 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.704010 (rank : 26) | θ value | 3.19293e-05 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.694095 (rank : 27) | θ value | 0.0193708 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.693307 (rank : 28) | θ value | 9.29e-05 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.691061 (rank : 29) | θ value | 7.59969e-07 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.685714 (rank : 30) | θ value | 4.92598e-06 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.683615 (rank : 31) | θ value | 7.59969e-07 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.683338 (rank : 32) | θ value | 0.00509761 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.682587 (rank : 33) | θ value | 6.43352e-06 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.682577 (rank : 34) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.682473 (rank : 35) | θ value | 6.43352e-06 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.682212 (rank : 36) | θ value | 6.43352e-06 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.677127 (rank : 37) | θ value | 2.44474e-05 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.675719 (rank : 38) | θ value | 3.19293e-05 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.671451 (rank : 39) | θ value | 0.0330416 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.667798 (rank : 40) | θ value | 0.125558 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.665931 (rank : 41) | θ value | 0.000270298 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.663885 (rank : 42) | θ value | 0.000602161 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.657938 (rank : 43) | θ value | 0.00102713 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.657553 (rank : 44) | θ value | 0.0148317 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.656789 (rank : 45) | θ value | 0.00102713 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.650113 (rank : 46) | θ value | 0.0193708 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.647736 (rank : 47) | θ value | 0.00228821 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.647566 (rank : 48) | θ value | 0.00228821 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.645027 (rank : 49) | θ value | 0.00228821 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.644262 (rank : 50) | θ value | 0.00134147 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.642142 (rank : 51) | θ value | 0.00509761 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.641162 (rank : 52) | θ value | 0.00134147 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.627167 (rank : 53) | θ value | 0.0736092 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.619965 (rank : 54) | θ value | 0.0431538 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.618654 (rank : 55) | θ value | 0.0431538 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.605703 (rank : 56) | θ value | 0.00175202 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.604026 (rank : 57) | θ value | 0.00665767 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.602178 (rank : 58) | θ value | 0.00228821 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.579030 (rank : 59) | θ value | 0.00035302 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.577404 (rank : 60) | θ value | 0.00035302 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.548700 (rank : 61) | θ value | 9.29e-05 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.546618 (rank : 62) | θ value | 9.29e-05 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.490673 (rank : 63) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.489358 (rank : 64) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.448207 (rank : 65) | θ value | 0.0961366 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.444553 (rank : 66) | θ value | 0.0961366 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.407394 (rank : 67) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
CU055_HUMAN
|
||||||
| NC score | 0.372618 (rank : 68) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.337304 (rank : 69) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.321893 (rank : 70) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.303395 (rank : 71) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
TIM14_HUMAN
|
||||||
| NC score | 0.100805 (rank : 72) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| NC score | 0.100323 (rank : 73) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
DCJ15_HUMAN
|
||||||
| NC score | 0.095588 (rank : 74) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.082631 (rank : 75) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.081542 (rank : 76) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.080710 (rank : 77) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
DCJ15_MOUSE
|
||||||
| NC score | 0.077018 (rank : 78) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.072594 (rank : 79) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.063437 (rank : 80) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.057258 (rank : 81) | θ value | 0.0252991 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.056602 (rank : 82) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.048143 (rank : 83) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.046759 (rank : 84) | θ value | 0.47712 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.044597 (rank : 85) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.042280 (rank : 86) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.038533 (rank : 87) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.037274 (rank : 88) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SPC25_MOUSE
|
||||||
| NC score | 0.037214 (rank : 89) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.036619 (rank : 90) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.035379 (rank : 91) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.035101 (rank : 92) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.033648 (rank : 93) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.033149 (rank : 94) | θ value | 0.163984 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.033134 (rank : 95) | θ value | 0.365318 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.032622 (rank : 96) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.032553 (rank : 97) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.032000 (rank : 98) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.031571 (rank : 99) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
TMCO1_HUMAN
|
||||||
| NC score | 0.031451 (rank : 100) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM00, O75545, Q9BZS3, Q9BZU8 | Gene names | TMCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 1 (Xenogeneic cross-immune protein PCIA3). | |||||
|
TMCO1_MOUSE
|
||||||
| NC score | 0.031451 (rank : 101) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921L3, Q3TS11 | Gene names | Tmco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 1. | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.031162 (rank : 102) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.031090 (rank : 103) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.030945 (rank : 104) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.029835 (rank : 105) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.029590 (rank : 106) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.029492 (rank : 107) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.029122 (rank : 108) | θ value | 0.21417 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.029017 (rank : 109) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.028660 (rank : 110) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.028383 (rank : 111) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.027266 (rank : 112) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.026943 (rank : 113) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.026481 (rank : 114) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.026165 (rank : 115) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ING4_HUMAN
|
||||||
| NC score | 0.025414 (rank : 116) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.025166 (rank : 117) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.024058 (rank : 118) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.024052 (rank : 119) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.023390 (rank : 120) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ING4_MOUSE
|
||||||
| NC score | 0.022492 (rank : 121) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.021720 (rank : 122) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.021403 (rank : 123) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.021017 (rank : 124) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.019688 (rank : 125) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.019347 (rank : 126) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
EXOSX_HUMAN
|
||||||
| NC score | 0.019274 (rank : 127) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.019177 (rank : 128) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ENPL_HUMAN
|
||||||
| NC score | 0.018515 (rank : 129) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.018198 (rank : 130) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.015969 (rank : 131) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.015923 (rank : 132) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.014946 (rank : 133) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.014438 (rank : 134) | θ value | 0.365318 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.014366 (rank : 135) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
KNG1_HUMAN
|
||||||
| NC score | 0.013249 (rank : 136) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01042, P01043 | Gene names | KNG1, BDK, KNG | |||
|
Domain Architecture |

|
|||||
| Description | Kininogen-1 precursor (Alpha-2-thiol proteinase inhibitor) [Contains: Kininogen-1 heavy chain; Bradykinin (Kallidin I); Lysyl-bradykinin (Kallidin II); Kininogen-1 light chain; Low molecular weight growth- promoting factor]. | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.012704 (rank : 137) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
K1718_MOUSE
|
||||||
| NC score | 0.012576 (rank : 138) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MCM8_HUMAN
|
||||||
| NC score | 0.012236 (rank : 139) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.012233 (rank : 140) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.011830 (rank : 141) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.011462 (rank : 142) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.011041 (rank : 143) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.010699 (rank : 144) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.010614 (rank : 145) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
PDCL2_MOUSE
|
||||||
| NC score | 0.010568 (rank : 146) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.009804 (rank : 147) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.009524 (rank : 148) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
K1C13_HUMAN
|
||||||
| NC score | 0.004873 (rank : 149) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.004843 (rank : 150) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.001301 (rank : 151) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.000646 (rank : 152) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | -0.001117 (rank : 153) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||