Please be patient as the page loads
|
DCJ14_HUMAN
|
||||||
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DCJ14_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992222 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 3) | NC score | 0.769258 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
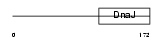
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 4) | NC score | 0.765798 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
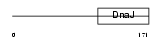
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DJC16_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 5) | NC score | 0.715805 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC16_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 6) | NC score | 0.715434 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 7) | NC score | 0.748530 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC18_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 8) | NC score | 0.737078 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 9) | NC score | 0.721374 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 10) | NC score | 0.687503 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 11) | NC score | 0.718839 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 12) | NC score | 0.718369 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 13) | NC score | 0.717183 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 14) | NC score | 0.737528 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 15) | NC score | 0.737383 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 16) | NC score | 0.706410 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 17) | NC score | 0.702181 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
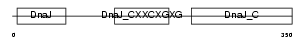
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 18) | NC score | 0.702357 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
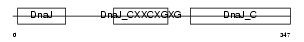
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 19) | NC score | 0.701248 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 20) | NC score | 0.703257 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 21) | NC score | 0.707643 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 22) | NC score | 0.727382 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 23) | NC score | 0.746180 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 24) | NC score | 0.706354 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 25) | NC score | 0.742961 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 26) | NC score | 0.693023 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 27) | NC score | 0.590395 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 28) | NC score | 0.586779 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 29) | NC score | 0.723974 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 30) | NC score | 0.703036 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 31) | NC score | 0.689699 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 32) | NC score | 0.701536 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 33) | NC score | 0.714062 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 34) | NC score | 0.714830 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 35) | NC score | 0.717907 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 36) | NC score | 0.710312 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
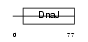
DNJC5_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 37) | NC score | 0.715139 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 38) | NC score | 0.714993 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
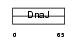
DNJB3_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 39) | NC score | 0.709051 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
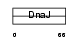
DNJB6_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 40) | NC score | 0.712041 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
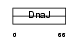
DNJB7_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 41) | NC score | 0.699359 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 42) | NC score | 0.681439 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 43) | NC score | 0.681674 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 44) | NC score | 0.706514 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
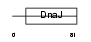
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 45) | NC score | 0.659476 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJ5B_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 46) | NC score | 0.702905 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
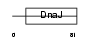
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 47) | NC score | 0.658251 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 48) | NC score | 0.541016 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 49) | NC score | 0.538371 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 50) | NC score | 0.688869 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
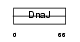
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 51) | NC score | 0.662979 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 52) | NC score | 0.565514 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 53) | NC score | 0.563535 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 54) | NC score | 0.653313 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 55) | NC score | 0.664293 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 56) | NC score | 0.622007 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 57) | NC score | 0.466552 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 58) | NC score | 0.466520 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
WBS18_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 59) | NC score | 0.664915 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 60) | NC score | 0.605703 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 61) | NC score | 0.673180 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 62) | NC score | 0.633769 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
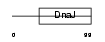
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DJC12_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 63) | NC score | 0.574358 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
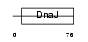
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
WBS18_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 64) | NC score | 0.649532 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 65) | NC score | 0.654833 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 66) | NC score | 0.631059 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
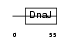
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 67) | NC score | 0.014971 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 68) | NC score | 0.025680 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 69) | NC score | 0.025136 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 70) | NC score | 0.025217 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.034499 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.029703 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.011337 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.011206 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.032558 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.025548 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.007772 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.008834 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.012967 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TIM14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.164214 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.163947 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.008261 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.002813 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
AUXI_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.096199 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.087300 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.009725 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
DCJ15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.156342 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
H2AW_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.016124 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.016316 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016314 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.017083 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.006616 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.015009 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.014846 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
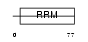
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.014636 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
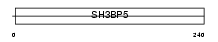
3BP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.013266 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |
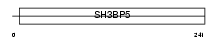
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.011110 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.019098 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.023162 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
DCJ15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.131221 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
GAK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.015824 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
SLBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.023306 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.004091 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | -0.000146 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
3BP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.011315 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
CHRD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.008756 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
H2AW_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.014924 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
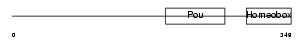
PO2F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.001998 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | -0.004039 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.007933 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
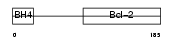
BCLX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.007004 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07817, Q5TE65, Q92976 | Gene names | BCL2L1, BCL2L, BCLX | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.001661 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
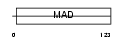
CT151_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.009611 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
SACS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.088552 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.089194 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SLBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.019692 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.312434 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
CU055_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.331886 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |
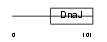
|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
CU055_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.359232 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
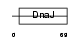
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.337881 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.332092 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.992222 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.769258 (rank : 3) | θ value | 8.38298e-14 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.765798 (rank : 4) | θ value | 1.86753e-13 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.748530 (rank : 5) | θ value | 6.64225e-11 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.746180 (rank : 6) | θ value | 4.76016e-09 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.742961 (rank : 7) | θ value | 8.11959e-09 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.737528 (rank : 8) | θ value | 4.30538e-10 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.737383 (rank : 9) | θ value | 4.30538e-10 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.737078 (rank : 10) | θ value | 8.67504e-11 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.727382 (rank : 11) | θ value | 4.76016e-09 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.723974 (rank : 12) | θ value | 1.80886e-08 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.721374 (rank : 13) | θ value | 1.133e-10 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.718839 (rank : 14) | θ value | 2.52405e-10 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.718369 (rank : 15) | θ value | 3.29651e-10 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.717907 (rank : 16) | θ value | 1.53129e-07 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.717183 (rank : 17) | θ value | 4.30538e-10 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.715805 (rank : 18) | θ value | 3.89403e-11 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.715434 (rank : 19) | θ value | 5.08577e-11 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.715139 (rank : 20) | θ value | 2.61198e-07 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.714993 (rank : 21) | θ value | 2.61198e-07 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.714830 (rank : 22) | θ value | 6.87365e-08 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.714062 (rank : 23) | θ value | 6.87365e-08 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.712041 (rank : 24) | θ value | 7.59969e-07 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.710312 (rank : 25) | θ value | 1.99992e-07 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.709051 (rank : 26) | θ value | 4.45536e-07 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.707643 (rank : 27) | θ value | 4.76016e-09 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.706514 (rank : 28) | θ value | 6.43352e-06 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.706410 (rank : 29) | θ value | 7.34386e-10 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.706354 (rank : 30) | θ value | 6.21693e-09 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.703257 (rank : 31) | θ value | 3.64472e-09 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.703036 (rank : 32) | θ value | 4.0297e-08 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.702905 (rank : 33) | θ value | 1.09739e-05 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.702357 (rank : 34) | θ value | 3.64472e-09 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
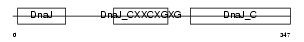
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.702181 (rank : 35) | θ value | 3.64472e-09 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
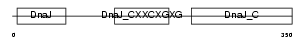
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.701536 (rank : 36) | θ value | 5.26297e-08 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.701248 (rank : 37) | θ value | 3.64472e-09 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.699359 (rank : 38) | θ value | 1.29631e-06 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.693023 (rank : 39) | θ value | 1.06045e-08 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.689699 (rank : 40) | θ value | 5.26297e-08 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.688869 (rank : 41) | θ value | 3.19293e-05 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.687503 (rank : 42) | θ value | 1.9326e-10 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.681674 (rank : 43) | θ value | 2.21117e-06 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.681439 (rank : 44) | θ value | 1.69304e-06 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.673180 (rank : 45) | θ value | 0.00175202 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.664915 (rank : 46) | θ value | 0.00134147 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.664293 (rank : 47) | θ value | 0.00035302 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.662979 (rank : 48) | θ value | 3.19293e-05 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.659476 (rank : 49) | θ value | 1.09739e-05 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.658251 (rank : 50) | θ value | 1.43324e-05 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.654833 (rank : 51) | θ value | 0.0193708 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.653313 (rank : 52) | θ value | 0.000270298 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.649532 (rank : 53) | θ value | 0.00869519 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.633769 (rank : 54) | θ value | 0.00298849 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.631059 (rank : 55) | θ value | 0.0252991 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.622007 (rank : 56) | θ value | 0.000602161 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.605703 (rank : 57) | θ value | 0.00175202 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.590395 (rank : 58) | θ value | 1.06045e-08 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.586779 (rank : 59) | θ value | 1.06045e-08 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.574358 (rank : 60) | θ value | 0.00665767 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.565514 (rank : 61) | θ value | 0.000270298 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.563535 (rank : 62) | θ value | 0.000270298 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.541016 (rank : 63) | θ value | 1.87187e-05 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.538371 (rank : 64) | θ value | 1.87187e-05 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.466552 (rank : 65) | θ value | 0.000786445 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.466520 (rank : 66) | θ value | 0.000786445 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.359232 (rank : 67) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.337881 (rank : 68) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.332092 (rank : 69) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CU055_HUMAN
|
||||||
| NC score | 0.331886 (rank : 70) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.312434 (rank : 71) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
TIM14_HUMAN
|
||||||
| NC score | 0.164214 (rank : 72) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| NC score | 0.163947 (rank : 73) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
DCJ15_HUMAN
|
||||||
| NC score | 0.156342 (rank : 74) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
DCJ15_MOUSE
|
||||||
| NC score | 0.131221 (rank : 75) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.096199 (rank : 76) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.089194 (rank : 77) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.088552 (rank : 78) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.087300 (rank : 79) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.034499 (rank : 80) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.032558 (rank : 81) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.029703 (rank : 82) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.025680 (rank : 83) | θ value | 0.163984 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.025548 (rank : 84) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.025217 (rank : 85) | θ value | 0.279714 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.025136 (rank : 86) | θ value | 0.21417 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
SLBP_MOUSE
|
||||||
| NC score | 0.023306 (rank : 87) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.023162 (rank : 88) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SLBP_HUMAN
|
||||||
| NC score | 0.019692 (rank : 89) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.019098 (rank : 90) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.017083 (rank : 91) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
H2AY_HUMAN
|
||||||
| NC score | 0.016316 (rank : 92) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| NC score | 0.016314 (rank : 93) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
H2AW_MOUSE
|
||||||
| NC score | 0.016124 (rank : 94) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.015824 (rank : 95) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.015009 (rank : 96) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.014971 (rank : 97) | θ value | 0.0961366 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
H2AW_HUMAN
|
||||||
| NC score | 0.014924 (rank : 98) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.014846 (rank : 99) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.014636 (rank : 100) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
3BP5_MOUSE
|
||||||
| NC score | 0.013266 (rank : 101) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.012967 (rank : 102) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.011337 (rank : 103) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.011315 (rank : 104) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.011206 (rank : 105) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.011110 (rank : 106) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.009725 (rank : 107) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.009611 (rank : 108) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.008834 (rank : 109) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CHRD_MOUSE
|
||||||
| NC score | 0.008756 (rank : 110) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.008261 (rank : 111) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.007933 (rank : 112) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.007772 (rank : 113) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
BCLX_HUMAN
|
||||||
| NC score | 0.007004 (rank : 114) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07817, Q5TE65, Q92976 | Gene names | BCL2L1, BCL2L, BCLX | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.006616 (rank : 115) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.004091 (rank : 116) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.002813 (rank : 117) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.001998 (rank : 118) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.001661 (rank : 119) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
TPR_HUMAN
|
||||||
| NC score | -0.000146 (rank : 120) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
WNK3_HUMAN
|
||||||
| NC score | -0.004039 (rank : 121) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||