Please be patient as the page loads
|
H2AW_HUMAN
|
||||||
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
H2AW_HUMAN
|
||||||
| θ value | 3.82926e-184 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| θ value | 7.22167e-183 (rank : 2) | NC score | 0.999678 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AY_HUMAN
|
||||||
| θ value | 3.07662e-133 (rank : 3) | NC score | 0.990268 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| θ value | 1.16911e-132 (rank : 4) | NC score | 0.990155 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
H2A2B_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 5) | NC score | 0.944077 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-B (H2a-613A). | |||||
|
H2A2B_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 6) | NC score | 0.943538 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUE6 | Gene names | HIST2H2AB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-B. | |||||
|
H2A1A_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 7) | NC score | 0.943618 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96QV6 | Gene names | HIST1H2AA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-A. | |||||
|
H2AX_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 8) | NC score | 0.945161 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16104, Q4ZGJ7, Q6IAS5 | Gene names | H2AFX, H2AX | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A.x (H2a/x). | |||||
|
H2AX_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 9) | NC score | 0.945564 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P27661 | Gene names | H2afx, H2a.x, H2ax, Hist5-2ax | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A.x (H2a/x). | |||||
|
H2A2A_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 10) | NC score | 0.941540 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6FI13, P20670 | Gene names | HIST2H2AA3, H2AFO, HIST2H2AA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-A (H2A.2). | |||||
|
H2A2A_MOUSE
|
||||||
| θ value | 2.96089e-35 (rank : 11) | NC score | 0.941540 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6GSS7, P20670, Q811M3 | Gene names | Hist2h2aa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-A (H2A.2) (H2a-614) (H2a-615). | |||||
|
H2A1K_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 12) | NC score | 0.941693 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP7 | Gene names | Hist1h2ak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-K. | |||||
|
H2A2C_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 13) | NC score | 0.942727 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16777, Q6DRA7, Q8IUE5 | Gene names | HIST2H2AC, H2AFQ | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 2-C (H2A-GL101) (H2A/r). | |||||
|
H2A2C_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 14) | NC score | 0.942727 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64523, Q8CGP3 | Gene names | Hist2h2ac, Hist2h2ab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-C (H2a-613B). | |||||
|
H2A3_HUMAN
|
||||||
| θ value | 5.0505e-35 (rank : 15) | NC score | 0.941252 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7L7L0 | Gene names | HIST3H2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 3. | |||||
|
H2A3_MOUSE
|
||||||
| θ value | 5.0505e-35 (rank : 16) | NC score | 0.941252 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BFU2 | Gene names | Hist3h2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 3. | |||||
|
H2A1C_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 17) | NC score | 0.940980 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93077, O00775, O00776, O00777, O00778, Q540R1 | Gene names | HIST1H2AC, H2AFL | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-C. | |||||
|
H2A1D_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 18) | NC score | 0.941396 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20671, P57754, Q6FGY6 | Gene names | HIST1H2AD, H2AFG | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-D (H2A.3). | |||||
|
H2A1E_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 19) | NC score | 0.941383 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28001, Q76P63 | Gene names | HIST1H2AE, H2AFA | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-E (H2A.2) (H2A/m). | |||||
|
H2A1H_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 20) | NC score | 0.942009 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP6 | Gene names | Hist1h2ah | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-H. | |||||
|
H2A1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 21) | NC score | 0.941383 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22752, P10812, Q5SZZ2 | Gene names | Hist1h2ab | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1. | |||||
|
H2A1H_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 22) | NC score | 0.941727 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KK5 | Gene names | HIST1H2AH, HIST1H2AI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-H. | |||||
|
H2A1J_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 23) | NC score | 0.941947 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99878, Q5JXQ5 | Gene names | HIST1H2AJ, H2AFE | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-J. | |||||
|
H2A1_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 24) | NC score | 0.941108 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P0C0S8, P02261, Q2M1R2, Q76PA6 | Gene names | HIST1H2AG, H2AFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1 (H2A.1). | |||||
|
H2A1F_MOUSE
|
||||||
| θ value | 1.9192e-34 (rank : 25) | NC score | 0.941497 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP5 | Gene names | Hist1h2af | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-F. | |||||
|
H2A1B_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 26) | NC score | 0.941536 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04908 | Gene names | HIST1H2AB, H2AFM | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-B. | |||||
|
H2AZ_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 27) | NC score | 0.938815 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P0C0S5, P17317, Q6I9U0 | Gene names | H2AFZ, H2AZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A.Z (H2A/z). | |||||
|
H2AZ_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 28) | NC score | 0.938815 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P0C0S6, P17317, Q3UA55, Q3UK43, Q68FD2 | Gene names | H2afz, H2az | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A.Z (H2A/z). | |||||
|
H2AV_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 29) | NC score | 0.938511 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q71UI9, Q59GV8, Q6PK98 | Gene names | H2AFV, H2AV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2AV (H2A.F/Z). | |||||
|
H2AV_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 30) | NC score | 0.938511 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3THW5, Q5NC92, Q6ZWU0 | Gene names | H2afv, H2av | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2AV (H2A.F/Z). | |||||
|
H2AFB_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 31) | NC score | 0.922552 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P98176, Q5TZB2, Q6FG78, Q96PR7 | Gene names | H2AFB3, H2ABBD, H2AFB | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A-Bbd (H2A Barr body-deficient) (H2A.Bbd). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 32) | NC score | 0.244232 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR14_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 33) | NC score | 0.239148 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
PARP9_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 34) | NC score | 0.251052 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |
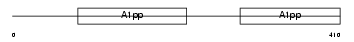
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
PARP9_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 35) | NC score | 0.228697 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |
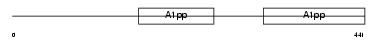
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PAR15_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 36) | NC score | 0.223998 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 37) | NC score | 0.298105 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 38) | NC score | 0.306746 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.250929 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.016387 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
ABTB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.062517 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.057652 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.001605 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.000780 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.008759 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.013449 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
PITX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.001934 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |
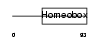
|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.007447 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.000247 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.014924 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.007261 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.004365 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.004816 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.006888 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | -0.005115 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
RPN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.010014 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBG6, Q3TKY0, Q91XH0 | Gene names | Rpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (EC 2.4.1.119) (Ribophorin II) (RPN-II). | |||||
|
DRAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.168593 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
DRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.163034 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
LRP16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.159847 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |
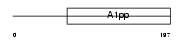
|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.158148 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |
|
|||||
| Description | Protein LRP16. | |||||
|
PAR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.069307 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056259 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.057357 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.061025 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.061061 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054960 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
H2AW_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.82926e-184 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| NC score | 0.999678 (rank : 2) | θ value | 7.22167e-183 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AY_HUMAN
|
||||||
| NC score | 0.990268 (rank : 3) | θ value | 3.07662e-133 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| NC score | 0.990155 (rank : 4) | θ value | 1.16911e-132 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
H2AX_MOUSE
|
||||||
| NC score | 0.945564 (rank : 5) | θ value | 2.26708e-35 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P27661 | Gene names | H2afx, H2a.x, H2ax, Hist5-2ax | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A.x (H2a/x). | |||||
|
H2AX_HUMAN
|
||||||
| NC score | 0.945161 (rank : 6) | θ value | 2.26708e-35 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16104, Q4ZGJ7, Q6IAS5 | Gene names | H2AFX, H2AX | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A.x (H2a/x). | |||||
|
H2A2B_MOUSE
|
||||||
| NC score | 0.944077 (rank : 7) | θ value | 4.568e-36 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-B (H2a-613A). | |||||
|
H2A1A_HUMAN
|
||||||
| NC score | 0.943618 (rank : 8) | θ value | 1.32909e-35 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96QV6 | Gene names | HIST1H2AA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-A. | |||||
|
H2A2B_HUMAN
|
||||||
| NC score | 0.943538 (rank : 9) | θ value | 1.01765e-35 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUE6 | Gene names | HIST2H2AB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-B. | |||||
|
H2A2C_HUMAN
|
||||||
| NC score | 0.942727 (rank : 10) | θ value | 3.86705e-35 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16777, Q6DRA7, Q8IUE5 | Gene names | HIST2H2AC, H2AFQ | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 2-C (H2A-GL101) (H2A/r). | |||||
|
H2A2C_MOUSE
|
||||||
| NC score | 0.942727 (rank : 11) | θ value | 3.86705e-35 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64523, Q8CGP3 | Gene names | Hist2h2ac, Hist2h2ab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-C (H2a-613B). | |||||
|
H2A1H_MOUSE
|
||||||
| NC score | 0.942009 (rank : 12) | θ value | 1.12514e-34 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP6 | Gene names | Hist1h2ah | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-H. | |||||
|
H2A1J_HUMAN
|
||||||
| NC score | 0.941947 (rank : 13) | θ value | 1.46948e-34 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99878, Q5JXQ5 | Gene names | HIST1H2AJ, H2AFE | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-J. | |||||
|
H2A1H_HUMAN
|
||||||
| NC score | 0.941727 (rank : 14) | θ value | 1.46948e-34 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KK5 | Gene names | HIST1H2AH, HIST1H2AI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-H. | |||||
|
H2A1K_MOUSE
|
||||||
| NC score | 0.941693 (rank : 15) | θ value | 3.86705e-35 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP7 | Gene names | Hist1h2ak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-K. | |||||
|
H2A2A_HUMAN
|
||||||
| NC score | 0.941540 (rank : 16) | θ value | 2.96089e-35 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6FI13, P20670 | Gene names | HIST2H2AA3, H2AFO, HIST2H2AA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-A (H2A.2). | |||||
|
H2A2A_MOUSE
|
||||||
| NC score | 0.941540 (rank : 17) | θ value | 2.96089e-35 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6GSS7, P20670, Q811M3 | Gene names | Hist2h2aa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 2-A (H2A.2) (H2a-614) (H2a-615). | |||||
|
H2A1B_HUMAN
|
||||||
| NC score | 0.941536 (rank : 18) | θ value | 9.52487e-34 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04908 | Gene names | HIST1H2AB, H2AFM | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-B. | |||||
|
H2A1F_MOUSE
|
||||||
| NC score | 0.941497 (rank : 19) | θ value | 1.9192e-34 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGP5 | Gene names | Hist1h2af | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1-F. | |||||
|
H2A1D_HUMAN
|
||||||
| NC score | 0.941396 (rank : 20) | θ value | 1.12514e-34 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20671, P57754, Q6FGY6 | Gene names | HIST1H2AD, H2AFG | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-D (H2A.3). | |||||
|
H2A1E_HUMAN
|
||||||
| NC score | 0.941383 (rank : 21) | θ value | 1.12514e-34 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28001, Q76P63 | Gene names | HIST1H2AE, H2AFA | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-E (H2A.2) (H2A/m). | |||||
|
H2A1_MOUSE
|
||||||
| NC score | 0.941383 (rank : 22) | θ value | 1.12514e-34 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22752, P10812, Q5SZZ2 | Gene names | Hist1h2ab | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1. | |||||
|
H2A3_HUMAN
|
||||||
| NC score | 0.941252 (rank : 23) | θ value | 5.0505e-35 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7L7L0 | Gene names | HIST3H2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 3. | |||||
|
H2A3_MOUSE
|
||||||
| NC score | 0.941252 (rank : 24) | θ value | 5.0505e-35 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BFU2 | Gene names | Hist3h2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 3. | |||||
|
H2A1_HUMAN
|
||||||
| NC score | 0.941108 (rank : 25) | θ value | 1.46948e-34 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P0C0S8, P02261, Q2M1R2, Q76PA6 | Gene names | HIST1H2AG, H2AFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A type 1 (H2A.1). | |||||
|
H2A1C_HUMAN
|
||||||
| NC score | 0.940980 (rank : 26) | θ value | 6.59618e-35 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93077, O00775, O00776, O00777, O00778, Q540R1 | Gene names | HIST1H2AC, H2AFL | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A type 1-C. | |||||
|
H2AZ_HUMAN
|
||||||
| NC score | 0.938815 (rank : 27) | θ value | 1.73987e-27 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P0C0S5, P17317, Q6I9U0 | Gene names | H2AFZ, H2AZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A.Z (H2A/z). | |||||
|
H2AZ_MOUSE
|
||||||
| NC score | 0.938815 (rank : 28) | θ value | 1.73987e-27 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P0C0S6, P17317, Q3UA55, Q3UK43, Q68FD2 | Gene names | H2afz, H2az | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2A.Z (H2A/z). | |||||
|
H2AV_HUMAN
|
||||||
| NC score | 0.938511 (rank : 29) | θ value | 2.96777e-27 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q71UI9, Q59GV8, Q6PK98 | Gene names | H2AFV, H2AV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2AV (H2A.F/Z). | |||||
|
H2AV_MOUSE
|
||||||
| NC score | 0.938511 (rank : 30) | θ value | 2.96777e-27 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3THW5, Q5NC92, Q6ZWU0 | Gene names | H2afv, H2av | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2AV (H2A.F/Z). | |||||
|
H2AFB_HUMAN
|
||||||
| NC score | 0.922552 (rank : 31) | θ value | 7.09661e-13 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P98176, Q5TZB2, Q6FG78, Q96PR7 | Gene names | H2AFB3, H2ABBD, H2AFB | |||
|
Domain Architecture |

|
|||||
| Description | Histone H2A-Bbd (H2A Barr body-deficient) (H2A.Bbd). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.306746 (rank : 32) | θ value | 0.00102713 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.298105 (rank : 33) | θ value | 0.000602161 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
PARP9_MOUSE
|
||||||
| NC score | 0.251052 (rank : 34) | θ value | 8.97725e-08 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.250929 (rank : 35) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.244232 (rank : 36) | θ value | 8.67504e-11 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR14_HUMAN
|
||||||
| NC score | 0.239148 (rank : 37) | θ value | 2.79066e-09 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
PARP9_HUMAN
|
||||||
| NC score | 0.228697 (rank : 38) | θ value | 2.88788e-06 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PAR15_HUMAN
|
||||||
| NC score | 0.223998 (rank : 39) | θ value | 8.40245e-06 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
DRAP1_HUMAN
|
||||||
| NC score | 0.168593 (rank : 40) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
DRAP1_MOUSE
|
||||||
| NC score | 0.163034 (rank : 41) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
LRP16_HUMAN
|
||||||
| NC score | 0.159847 (rank : 42) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| NC score | 0.158148 (rank : 43) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.069307 (rank : 44) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
ABTB2_MOUSE
|
||||||
| NC score | 0.062517 (rank : 45) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.061061 (rank : 46) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.061025 (rank : 47) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.057652 (rank : 48) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.057357 (rank : 49) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.056259 (rank : 50) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.054960 (rank : 51) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.016387 (rank : 52) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.014924 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.013449 (rank : 54) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
RPN2_MOUSE
|
||||||
| NC score | 0.010014 (rank : 55) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBG6, Q3TKY0, Q91XH0 | Gene names | Rpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (EC 2.4.1.119) (Ribophorin II) (RPN-II). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.008759 (rank : 56) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.007447 (rank : 57) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.007261 (rank : 58) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.006888 (rank : 59) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.004816 (rank : 60) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.004365 (rank : 61) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PITX2_MOUSE
|
||||||
| NC score | 0.001934 (rank : 62) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.001605 (rank : 63) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.000780 (rank : 64) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.000247 (rank : 65) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | -0.005115 (rank : 66) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||