Please be patient as the page loads
|
PARPT_MOUSE
|
||||||
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARPT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994878 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 4.11246e-53 (rank : 3) | NC score | 0.853098 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 9.48078e-50 (rank : 4) | NC score | 0.849420 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR15_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 5) | NC score | 0.790190 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
PAR14_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 6) | NC score | 0.707876 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 7) | NC score | 0.794226 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 6.38894e-30 (rank : 8) | NC score | 0.699786 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 9) | NC score | 0.697420 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PARP9_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 10) | NC score | 0.555220 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
PARP9_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 11) | NC score | 0.574351 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 12) | NC score | 0.145314 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 13) | NC score | 0.122517 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
PARP3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.153664 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.047123 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.106001 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.102668 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.025363 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
PARP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.090260 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.030859 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.025532 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.073183 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.024814 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.022504 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.074328 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PARP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.081556 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.042584 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.012926 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.080700 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
E2AK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.002258 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
KS6C1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.004589 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.015860 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.070769 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.002482 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LHX5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.005587 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |
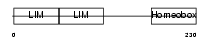
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
LHX5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.005585 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |
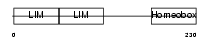
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.010786 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.017685 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.011723 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.013536 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.005323 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.041788 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
CHL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.001317 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
K1383_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.016071 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
S38A3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.008846 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCP2, Q8BS53, Q9JLL8 | Gene names | Slc38a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | System N amino acid transporter 1 (SN1) (N-system amino acid transporter 1) (Solute carrier family 38 member 3) (mNAT). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.009731 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.000672 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.052068 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
H2AW_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.061061 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.059777 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.077516 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.077152 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
LRP16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.173445 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |
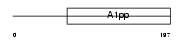
|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.166741 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |
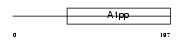
|
|||||
| Description | Protein LRP16. | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.994878 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.853098 (rank : 3) | θ value | 4.11246e-53 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.849420 (rank : 4) | θ value | 9.48078e-50 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.794226 (rank : 5) | θ value | 2.86786e-30 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
PAR15_HUMAN
|
||||||
| NC score | 0.790190 (rank : 6) | θ value | 1.6247e-33 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
PAR14_HUMAN
|
||||||
| NC score | 0.707876 (rank : 7) | θ value | 2.19584e-30 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.699786 (rank : 8) | θ value | 6.38894e-30 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.697420 (rank : 9) | θ value | 1.05554e-24 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PARP9_HUMAN
|
||||||
| NC score | 0.574351 (rank : 10) | θ value | 7.84624e-12 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PARP9_MOUSE
|
||||||
| NC score | 0.555220 (rank : 11) | θ value | 2.69671e-12 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
LRP16_HUMAN
|
||||||
| NC score | 0.173445 (rank : 12) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| NC score | 0.166741 (rank : 13) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.153664 (rank : 14) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.145314 (rank : 15) | θ value | 6.87365e-08 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.122517 (rank : 16) | θ value | 1.87187e-05 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.106001 (rank : 17) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.102668 (rank : 18) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
PARP2_MOUSE
|
||||||
| NC score | 0.090260 (rank : 19) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP1_HUMAN
|
||||||
| NC score | 0.081556 (rank : 20) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.080700 (rank : 21) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
H2AY_HUMAN
|
||||||
| NC score | 0.077516 (rank : 22) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| NC score | 0.077152 (rank : 23) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.074328 (rank : 24) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.073183 (rank : 25) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.070769 (rank : 26) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
H2AW_HUMAN
|
||||||
| NC score | 0.061061 (rank : 27) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| NC score | 0.059777 (rank : 28) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.052068 (rank : 29) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.047123 (rank : 30) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.042584 (rank : 31) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.041788 (rank : 32) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.030859 (rank : 33) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
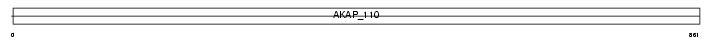
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.025532 (rank : 34) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.025363 (rank : 35) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.024814 (rank : 36) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.022504 (rank : 37) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.017685 (rank : 38) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
K1383_HUMAN
|
||||||
| NC score | 0.016071 (rank : 39) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.015860 (rank : 40) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.013536 (rank : 41) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.012926 (rank : 42) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.011723 (rank : 43) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.010786 (rank : 44) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.009731 (rank : 45) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
S38A3_MOUSE
|
||||||
| NC score | 0.008846 (rank : 46) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCP2, Q8BS53, Q9JLL8 | Gene names | Slc38a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | System N amino acid transporter 1 (SN1) (N-system amino acid transporter 1) (Solute carrier family 38 member 3) (mNAT). | |||||
|
LHX5_HUMAN
|
||||||
| NC score | 0.005587 (rank : 47) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
LHX5_MOUSE
|
||||||
| NC score | 0.005585 (rank : 48) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.005323 (rank : 49) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
KS6C1_HUMAN
|
||||||
| NC score | 0.004589 (rank : 50) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.002482 (rank : 51) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
E2AK4_HUMAN
|
||||||
| NC score | 0.002258 (rank : 52) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
CHL1_MOUSE
|
||||||
| NC score | 0.001317 (rank : 53) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.000672 (rank : 54) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||