Please be patient as the page loads
|
DHX57_HUMAN
|
||||||
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX57_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993141 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 6.54689e-176 (rank : 3) | NC score | 0.960686 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 1.97121e-172 (rank : 4) | NC score | 0.963242 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 1.56551e-161 (rank : 5) | NC score | 0.958820 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 3.4876e-161 (rank : 6) | NC score | 0.954361 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 1.02419e-128 (rank : 7) | NC score | 0.951104 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 2.78914e-126 (rank : 8) | NC score | 0.949779 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 5.91091e-68 (rank : 9) | NC score | 0.921915 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 10) | NC score | 0.921172 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 11) | NC score | 0.887866 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 7.24236e-58 (rank : 12) | NC score | 0.894962 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 1.46268e-50 (rank : 13) | NC score | 0.834928 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 3.2585e-50 (rank : 14) | NC score | 0.868734 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 15) | NC score | 0.909873 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 5.7518e-47 (rank : 16) | NC score | 0.867369 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 17) | NC score | 0.879405 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 18) | NC score | 0.879218 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 19) | NC score | 0.879516 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 20) | NC score | 0.869452 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 21) | NC score | 0.865140 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 22) | NC score | 0.862484 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 2.05049e-36 (rank : 23) | NC score | 0.861700 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 24) | NC score | 0.838629 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.077139 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.093384 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
NUPL2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 27) | NC score | 0.071790 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 28) | NC score | 0.031939 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.082412 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 30) | NC score | 0.082412 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 31) | NC score | 0.018176 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 32) | NC score | 0.072112 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 33) | NC score | 0.071560 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 34) | NC score | 0.067839 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.085027 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.076147 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.058349 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.055286 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.047256 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.047123 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.052535 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.062333 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.079786 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.079007 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.071118 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.013274 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.004945 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
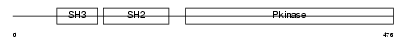
SRMS_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | -0.001806 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.067180 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
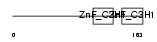
TTP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.059232 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.061680 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.065460 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.038846 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.060962 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.041321 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.032503 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
NU107_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.034331 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.046796 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
XPO6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.017551 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QU8, Q2YDX3, Q53G88, Q68G50, Q76N88, Q96CP8, Q9BT21 | Gene names | XPO6, KIAA0370, RANBP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.064927 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.066773 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.066121 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.074984 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.050164 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.022089 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.065193 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.011660 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
NU107_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.031241 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
NUB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.034422 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.057297 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.056280 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.033690 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.029011 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.029562 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
NUB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.032193 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54729, Q8K3U0 | Gene names | Nub1, Nyren18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (Protein BS4). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.019678 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.004360 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.006855 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.004388 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.038590 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
TRI16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.006967 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
KYNU_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.012302 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXF0 | Gene names | Kynu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kynureninase (EC 3.7.1.3) (L-kynurenine hydrolase). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.003376 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
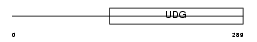
TDG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.012771 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |
|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
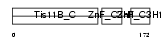
TISB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.037112 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
APEH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.008633 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R146, Q8K029, Q8R0M9 | Gene names | Apeh | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.002896 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.006527 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.033691 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.024073 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CCD60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.009087 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWA6 | Gene names | CCDC60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
CROP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.006763 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.006801 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
CX040_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.009438 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1F3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf40 homolog. | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.021565 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
PYGM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.004400 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.032076 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
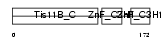
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.005244 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRI16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.005440 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PP9, Q7TPY6, Q8C332, Q8C6V0 | Gene names | Trim16, Ebbp | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
TRM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.014415 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.017010 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.002414 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.004799 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.000651 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
MBN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.028807 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.001907 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.000607 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
SDK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | -0.001050 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | -0.001948 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.993141 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.963242 (rank : 3) | θ value | 1.97121e-172 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.960686 (rank : 4) | θ value | 6.54689e-176 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.958820 (rank : 5) | θ value | 1.56551e-161 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.954361 (rank : 6) | θ value | 3.4876e-161 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.951104 (rank : 7) | θ value | 1.02419e-128 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.949779 (rank : 8) | θ value | 2.78914e-126 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.921915 (rank : 9) | θ value | 5.91091e-68 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.921172 (rank : 10) | θ value | 1.11475e-66 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.909873 (rank : 11) | θ value | 1.51363e-47 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.894962 (rank : 12) | θ value | 7.24236e-58 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.887866 (rank : 13) | θ value | 4.53632e-60 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.879516 (rank : 14) | θ value | 1.01529e-43 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.879405 (rank : 15) | θ value | 1.08474e-45 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.879218 (rank : 16) | θ value | 1.08474e-45 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.869452 (rank : 17) | θ value | 5.57106e-42 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.868734 (rank : 18) | θ value | 3.2585e-50 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.867369 (rank : 19) | θ value | 5.7518e-47 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.865140 (rank : 20) | θ value | 2.76489e-41 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.862484 (rank : 21) | θ value | 1.57e-36 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.861700 (rank : 22) | θ value | 2.05049e-36 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.838629 (rank : 23) | θ value | 1.01765e-35 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.834928 (rank : 24) | θ value | 1.46268e-50 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.093384 (rank : 25) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.085027 (rank : 26) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.082412 (rank : 27) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.082412 (rank : 28) | θ value | 0.0193708 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.079786 (rank : 29) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.079007 (rank : 30) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.077139 (rank : 31) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.076147 (rank : 32) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.074984 (rank : 33) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.072112 (rank : 34) | θ value | 0.0330416 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
NUPL2_HUMAN
|
||||||
| NC score | 0.071790 (rank : 35) | θ value | 0.0148317 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.071560 (rank : 36) | θ value | 0.0563607 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.071118 (rank : 37) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.067839 (rank : 38) | θ value | 0.0736092 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.067180 (rank : 39) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.066773 (rank : 40) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.066121 (rank : 41) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.065460 (rank : 42) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.065193 (rank : 43) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.064927 (rank : 44) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.062333 (rank : 45) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.061680 (rank : 46) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.060962 (rank : 47) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.059232 (rank : 48) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.058349 (rank : 49) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.057297 (rank : 50) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.056280 (rank : 51) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.055286 (rank : 52) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.052535 (rank : 53) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.050164 (rank : 54) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.047256 (rank : 55) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.047123 (rank : 56) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.046796 (rank : 57) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.041321 (rank : 58) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.038846 (rank : 59) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
PEO1_MOUSE
|
||||||
| NC score | 0.038590 (rank : 60) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.037112 (rank : 61) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
NUB1_HUMAN
|
||||||
| NC score | 0.034422 (rank : 62) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
NU107_HUMAN
|
||||||
| NC score | 0.034331 (rank : 63) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.033691 (rank : 64) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.033690 (rank : 65) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.032503 (rank : 66) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
NUB1_MOUSE
|
||||||
| NC score | 0.032193 (rank : 67) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54729, Q8K3U0 | Gene names | Nub1, Nyren18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (Protein BS4). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.032076 (rank : 68) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.031939 (rank : 69) | θ value | 0.0193708 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
NU107_MOUSE
|
||||||
| NC score | 0.031241 (rank : 70) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.029562 (rank : 71) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_HUMAN
|
||||||
| NC score | 0.029011 (rank : 72) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.028807 (rank : 73) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.024073 (rank : 74) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.022089 (rank : 75) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.021565 (rank : 76) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.019678 (rank : 77) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.018176 (rank : 78) | θ value | 0.0330416 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
XPO6_HUMAN
|
||||||
| NC score | 0.017551 (rank : 79) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QU8, Q2YDX3, Q53G88, Q68G50, Q76N88, Q96CP8, Q9BT21 | Gene names | XPO6, KIAA0370, RANBP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.017010 (rank : 80) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
TRM1_HUMAN
|
||||||
| NC score | 0.014415 (rank : 81) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.013274 (rank : 82) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
TDG_HUMAN
|
||||||
| NC score | 0.012771 (rank : 83) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |

|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
KYNU_MOUSE
|
||||||
| NC score | 0.012302 (rank : 84) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXF0 | Gene names | Kynu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kynureninase (EC 3.7.1.3) (L-kynurenine hydrolase). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.011660 (rank : 85) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CX040_MOUSE
|
||||||
| NC score | 0.009438 (rank : 86) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1F3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf40 homolog. | |||||
|
CCD60_HUMAN
|
||||||
| NC score | 0.009087 (rank : 87) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWA6 | Gene names | CCDC60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
APEH_MOUSE
|
||||||
| NC score | 0.008633 (rank : 88) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R146, Q8K029, Q8R0M9 | Gene names | Apeh | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase). | |||||
|
TRI16_HUMAN
|
||||||
| NC score | 0.006967 (rank : 89) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.006855 (rank : 90) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.006801 (rank : 91) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.006763 (rank : 92) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.006527 (rank : 93) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
TRI16_MOUSE
|
||||||
| NC score | 0.005440 (rank : 94) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PP9, Q7TPY6, Q8C332, Q8C6V0 | Gene names | Trim16, Ebbp | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.005244 (rank : 95) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.004945 (rank : 96) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.004799 (rank : 97) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
PYGM_MOUSE
|
||||||
| NC score | 0.004400 (rank : 98) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.004388 (rank : 99) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.004360 (rank : 100) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.003376 (rank : 101) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.002896 (rank : 102) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.002414 (rank : 103) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.001907 (rank : 104) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.000651 (rank : 105) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | -0.000607 (rank : 106) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
SDK2_HUMAN
|
||||||
| NC score | -0.001050 (rank : 107) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SRMS_HUMAN
|
||||||
| NC score | -0.001806 (rank : 108) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.001948 (rank : 109) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||