Please be patient as the page loads
|
DHX29_HUMAN
|
||||||
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX29_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991082 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 1.6163e-166 (rank : 3) | NC score | 0.959894 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 3.4876e-161 (rank : 4) | NC score | 0.954361 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 4.55495e-161 (rank : 5) | NC score | 0.954149 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 2.95243e-160 (rank : 6) | NC score | 0.964225 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 1.87085e-122 (rank : 7) | NC score | 0.950076 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 6.88586e-117 (rank : 8) | NC score | 0.949457 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 9) | NC score | 0.891835 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 1.90147e-66 (rank : 10) | NC score | 0.890379 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 11) | NC score | 0.900041 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 1.50663e-63 (rank : 12) | NC score | 0.899898 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 13) | NC score | 0.918843 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 1.66577e-62 (rank : 14) | NC score | 0.917887 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 15) | NC score | 0.885999 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 4.85792e-54 (rank : 16) | NC score | 0.900046 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 3.60269e-49 (rank : 17) | NC score | 0.879670 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 4.70527e-49 (rank : 18) | NC score | 0.880117 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 19) | NC score | 0.910141 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 20) | NC score | 0.892782 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 21) | NC score | 0.880265 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 22) | NC score | 0.850315 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 4.568e-36 (rank : 23) | NC score | 0.838151 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 24) | NC score | 0.877816 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.045246 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 26) | NC score | 0.053491 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.050220 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.039591 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.040537 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.011370 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.023525 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.040272 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.037997 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.059282 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.036094 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.065888 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.011929 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.030949 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.063227 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.027990 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.023641 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.017395 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SYS_HUMAN
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.030914 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49591, Q9NSE3 | Gene names | SARS, SERS | |||
|
Domain Architecture |
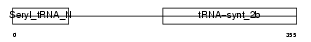
|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.029671 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DIRA3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.005630 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |
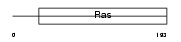
|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.025065 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.022099 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NBN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.024592 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.009591 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
LYAR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.040291 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.021524 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.027812 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.030848 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.032649 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.028246 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.017289 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.036155 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.025557 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
SYS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.029118 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26638 | Gene names | Sars, Sars1, Sers | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.036093 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.051462 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.029468 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.025514 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.009474 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.018140 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.027796 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.041466 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.019928 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PRPF3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.032257 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |
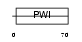
|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.032395 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.024390 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CENPK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.021124 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ESN5, Q3UXA9, Q569Q3, Q8C469 | Gene names | Cenpk, Solt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (SoxLZ/Sox6 leucine zipper-binding protein in testis). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.026312 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.024964 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.025088 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NIN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.024507 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.032990 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
ABC8A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.005268 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K442, Q6PAV3, Q8C0A9, Q8R0R4 | Gene names | Abca8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-A. | |||||
|
AL2SA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.019048 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.025379 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.039093 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.018497 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.017427 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.016882 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.025715 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.025772 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.030416 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.001033 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.031561 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.021741 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.004184 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.001689 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.026615 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.007979 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.025670 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.025687 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.015096 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CCD27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.025404 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.007171 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.000190 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.023271 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NOC2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.017688 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |
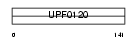
|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PI3R5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.013176 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SW28, Q3TU01, Q3UDZ2, Q8C215, Q8CGQ7 | Gene names | Pik3r5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit). | |||||
|
CCD45_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.022668 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96GE4, Q96M81 | Gene names | CCDC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.029990 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.029990 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.020374 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.008068 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.026225 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.024137 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.028899 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.016122 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
STMN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.017337 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |
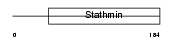
|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.017334 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.027580 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
A4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.011307 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.011536 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ABCAA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.003892 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWZ4, Q6PIQ6, Q7Z2I9, Q7Z7P7, Q86TD2 | Gene names | ABCA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 10. | |||||
|
ABCCB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.003416 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J66, Q8TDJ0, Q96JA6, Q9BX80 | Gene names | ABCC11, MRP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette transporter sub-family C member 11 (Multidrug resistance-associated protein 8). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.037772 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.038518 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.028055 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.028033 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
FTS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.008169 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64362 | Gene names | Fts, Fif, Ft1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein (FT1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.028587 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.009656 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
UTP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.012222 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYH9, Q8IX96, Q96BL2, Q9NQ91 | Gene names | UTP6, C17orf40, HCA66, MHAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 6 homolog (Multiple hat domains protein) (Hepatocellular carcinoma-associated antigen 66). | |||||
|
AMFR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.003308 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
BFSP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.007697 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
CP2CJ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.000932 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |
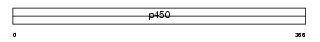
|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.026722 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.004165 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
FOXN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.001161 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |
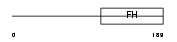
|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.020706 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
ITA11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.004034 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
MA2A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.004080 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27046 | Gene names | Man2a1, Mana2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1) (AMAN II). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.006651 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.008711 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.015962 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.017129 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PI3R5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.010885 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYR1, Q5G936, Q5G938, Q5G939, Q8IZ23, Q9Y2Y2 | Gene names | PIK3R5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit) (Protein FOAP-2). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.017458 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.016119 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.010351 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.991082 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.964225 (rank : 3) | θ value | 2.95243e-160 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.959894 (rank : 4) | θ value | 1.6163e-166 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.954361 (rank : 5) | θ value | 3.4876e-161 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.954149 (rank : 6) | θ value | 4.55495e-161 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.950076 (rank : 7) | θ value | 1.87085e-122 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.949457 (rank : 8) | θ value | 6.88586e-117 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.918843 (rank : 9) | θ value | 1.96772e-63 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.917887 (rank : 10) | θ value | 1.66577e-62 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.910141 (rank : 11) | θ value | 3.85808e-43 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.900046 (rank : 12) | θ value | 4.85792e-54 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.900041 (rank : 13) | θ value | 1.50663e-63 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.899898 (rank : 14) | θ value | 1.50663e-63 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.892782 (rank : 15) | θ value | 3.85808e-43 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.891835 (rank : 16) | θ value | 2.92676e-75 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.890379 (rank : 17) | θ value | 1.90147e-66 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.885999 (rank : 18) | θ value | 4.53632e-60 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.880265 (rank : 19) | θ value | 1.12253e-42 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.880117 (rank : 20) | θ value | 4.70527e-49 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.879670 (rank : 21) | θ value | 3.60269e-49 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.877816 (rank : 22) | θ value | 1.79631e-32 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.850315 (rank : 23) | θ value | 2.76489e-41 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.838151 (rank : 24) | θ value | 4.568e-36 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.065888 (rank : 25) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.063227 (rank : 26) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.059282 (rank : 27) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.053491 (rank : 28) | θ value | 0.0330416 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.051462 (rank : 29) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.050220 (rank : 30) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.045246 (rank : 31) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.041466 (rank : 32) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.040537 (rank : 33) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
LYAR_HUMAN
|
||||||
| NC score | 0.040291 (rank : 34) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.040272 (rank : 35) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.039591 (rank : 36) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.039093 (rank : 37) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.038518 (rank : 38) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.037997 (rank : 39) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.037772 (rank : 40) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.036155 (rank : 41) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.036094 (rank : 42) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.036093 (rank : 43) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.032990 (rank : 44) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.032649 (rank : 45) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.032395 (rank : 46) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
PRPF3_HUMAN
|
||||||
| NC score | 0.032257 (rank : 47) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.031561 (rank : 48) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.030949 (rank : 49) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYS_HUMAN
|
||||||
| NC score | 0.030914 (rank : 50) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49591, Q9NSE3 | Gene names | SARS, SERS | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.030848 (rank : 51) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.030416 (rank : 52) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.029990 (rank : 53) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.029990 (rank : 54) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.029671 (rank : 55) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.029468 (rank : 56) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
SYS_MOUSE
|
||||||
| NC score | 0.029118 (rank : 57) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26638 | Gene names | Sars, Sars1, Sers | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.028899 (rank : 58) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.028587 (rank : 59) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.028246 (rank : 60) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.028055 (rank : 61) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.028033 (rank : 62) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.027990 (rank : 63) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.027812 (rank : 64) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.027796 (rank : 65) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.027580 (rank : 66) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.026722 (rank : 67) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.026615 (rank : 68) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.026312 (rank : 69) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.026225 (rank : 70) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.025772 (rank : 71) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.025715 (rank : 72) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.025687 (rank : 73) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.025670 (rank : 74) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.025557 (rank : 75) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.025514 (rank : 76) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.025404 (rank : 77) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.025379 (rank : 78) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.025088 (rank : 79) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.025065 (rank : 80) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.024964 (rank : 81) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.024592 (rank : 82) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.024507 (rank : 83) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.024390 (rank : 84) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.024137 (rank : 85) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.023641 (rank : 86) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.023525 (rank : 87) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.023271 (rank : 88) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CCD45_HUMAN
|
||||||
| NC score | 0.022668 (rank : 89) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96GE4, Q96M81 | Gene names | CCDC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.022099 (rank : 90) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.021741 (rank : 91) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.021524 (rank : 92) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CENPK_MOUSE
|
||||||
| NC score | 0.021124 (rank : 93) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ESN5, Q3UXA9, Q569Q3, Q8C469 | Gene names | Cenpk, Solt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (SoxLZ/Sox6 leucine zipper-binding protein in testis). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.020706 (rank : 94) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.020374 (rank : 95) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.019928 (rank : 96) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.019048 (rank : 97) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.018497 (rank : 98) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.018140 (rank : 99) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NOC2L_MOUSE
|
||||||
| NC score | 0.017688 (rank : 100) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.017458 (rank : 101) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.017427 (rank : 102) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.017395 (rank : 103) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
STMN4_HUMAN
|
||||||
| NC score | 0.017337 (rank : 104) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| NC score | 0.017334 (rank : 105) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.017289 (rank : 106) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.017129 (rank : 107) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.016882 (rank : 108) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.016122 (rank : 109) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.016119 (rank : 110) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.015962 (rank : 111) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.015096 (rank : 112) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
PI3R5_MOUSE
|
||||||
| NC score | 0.013176 (rank : 113) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SW28, Q3TU01, Q3UDZ2, Q8C215, Q8CGQ7 | Gene names | Pik3r5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit). | |||||
|
UTP6_HUMAN
|
||||||
| NC score | 0.012222 (rank : 114) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYH9, Q8IX96, Q96BL2, Q9NQ91 | Gene names | UTP6, C17orf40, HCA66, MHAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 6 homolog (Multiple hat domains protein) (Hepatocellular carcinoma-associated antigen 66). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.011929 (rank : 115) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.011536 (rank : 116) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.011370 (rank : 117) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.011307 (rank : 118) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
PI3R5_HUMAN
|
||||||
| NC score | 0.010885 (rank : 119) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYR1, Q5G936, Q5G938, Q5G939, Q8IZ23, Q9Y2Y2 | Gene names | PIK3R5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 5 (PI3-kinase regulatory subunit 5) (PI3-kinase p101 subunit) (PtdIns-3-kinase p101) (p101- PI3K) (Phosphatidylinositol-4,5-bisphosphate 3-kinase regulatory subunit) (PtdIns-3-kinase regulatory subunit) (Protein FOAP-2). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.010351 (rank : 120) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.009656 (rank : 121) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.009591 (rank : 122) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.009474 (rank : 123) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.008711 (rank : 124) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
FTS1_MOUSE
|
||||||
| NC score | 0.008169 (rank : 125) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64362 | Gene names | Fts, Fif, Ft1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein (FT1). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.008068 (rank : 126) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.007979 (rank : 127) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
BFSP2_MOUSE
|
||||||
| NC score | 0.007697 (rank : 128) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.007171 (rank : 129) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.006651 (rank : 130) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
DIRA3_HUMAN
|
||||||
| NC score | 0.005630 (rank : 131) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
ABC8A_MOUSE
|
||||||
| NC score | 0.005268 (rank : 132) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K442, Q6PAV3, Q8C0A9, Q8R0R4 | Gene names | Abca8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-A. | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.004184 (rank : 133) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.004165 (rank : 134) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
MA2A1_MOUSE
|
||||||
| NC score | 0.004080 (rank : 135) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27046 | Gene names | Man2a1, Mana2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1) (AMAN II). | |||||
|
ITA11_HUMAN
|
||||||
| NC score | 0.004034 (rank : 136) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ABCAA_HUMAN
|
||||||
| NC score | 0.003892 (rank : 137) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWZ4, Q6PIQ6, Q7Z2I9, Q7Z7P7, Q86TD2 | Gene names | ABCA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 10. | |||||
|
ABCCB_HUMAN
|
||||||
| NC score | 0.003416 (rank : 138) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J66, Q8TDJ0, Q96JA6, Q9BX80 | Gene names | ABCC11, MRP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette transporter sub-family C member 11 (Multidrug resistance-associated protein 8). | |||||
|
AMFR2_HUMAN
|
||||||
| NC score | 0.003308 (rank : 139) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.001689 (rank : 140) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
FOXN3_HUMAN
|
||||||
| NC score | 0.001161 (rank : 141) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |

|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.001033 (rank : 142) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
CP2CJ_HUMAN
|
||||||
| NC score | 0.000932 (rank : 143) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.000190 (rank : 144) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||