Please be patient as the page loads
|
ITAM_HUMAN
|
||||||
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITAD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994244 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13349, Q15575, Q15576 | Gene names | ITGAD | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-D precursor (Leukointegrin alpha D) (ADB2) (CD11d antigen). | |||||
|
ITAD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995100 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3V0T4 | Gene names | Itgad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-D precursor (CD11d antigen). | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996836 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAX_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994212 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
ITAX_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.995859 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QXH4 | Gene names | Itgax | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (CD11c antigen). | |||||
|
ITAL_HUMAN
|
||||||
| θ value | 1.16101e-156 (rank : 7) | NC score | 0.977998 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
ITAL_MOUSE
|
||||||
| θ value | 2.18956e-155 (rank : 8) | NC score | 0.977744 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
ITAE_HUMAN
|
||||||
| θ value | 3.42535e-108 (rank : 9) | NC score | 0.962048 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P38570, Q9NZU9 | Gene names | ITGAE | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Mucosal lymphocyte 1 antigen) (HML-1 antigen) (Integrin alpha-IEL) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITAE_MOUSE
|
||||||
| θ value | 1.48926e-103 (rank : 10) | NC score | 0.961314 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60677 | Gene names | Itgae | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Integrin alpha M290) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA1_HUMAN
|
||||||
| θ value | 4.79078e-102 (rank : 11) | NC score | 0.952221 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P56199 | Gene names | ITGA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-1 (Laminin and collagen receptor) (VLA-1) (CD49a antigen). | |||||
|
ITA11_HUMAN
|
||||||
| θ value | 4.95767e-99 (rank : 12) | NC score | 0.950958 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITA10_HUMAN
|
||||||
| θ value | 1.22112e-97 (rank : 13) | NC score | 0.952443 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
ITA2_MOUSE
|
||||||
| θ value | 2.08291e-97 (rank : 14) | NC score | 0.947758 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62469, Q62163 | Gene names | Itga2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITA2_HUMAN
|
||||||
| θ value | 1.49272e-95 (rank : 15) | NC score | 0.946551 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17301, Q14595 | Gene names | ITGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITA11_MOUSE
|
||||||
| θ value | 8.19079e-94 (rank : 16) | NC score | 0.948418 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P61622 | Gene names | Itga11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITA4_MOUSE
|
||||||
| θ value | 1.66964e-54 (rank : 17) | NC score | 0.843711 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
ITA4_HUMAN
|
||||||
| θ value | 1.01294e-51 (rank : 18) | NC score | 0.839780 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
ITA9_HUMAN
|
||||||
| θ value | 1.91031e-50 (rank : 19) | NC score | 0.845771 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
ITA3_MOUSE
|
||||||
| θ value | 1.12253e-42 (rank : 20) | NC score | 0.805944 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA3_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 21) | NC score | 0.805001 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26006 | Gene names | ITGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (FRP-2) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITAV_MOUSE
|
||||||
| θ value | 8.32485e-38 (rank : 22) | NC score | 0.780182 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43406 | Gene names | Itgav | |||
|
Domain Architecture |
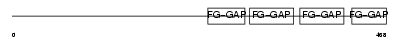
|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA5_MOUSE
|
||||||
| θ value | 1.85459e-37 (rank : 23) | NC score | 0.778618 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITAV_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 24) | NC score | 0.775737 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06756 | Gene names | ITGAV, VNRA | |||
|
Domain Architecture |
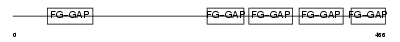
|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA5_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 25) | NC score | 0.777954 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |
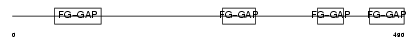
|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA6_MOUSE
|
||||||
| θ value | 3.27365e-34 (rank : 26) | NC score | 0.773461 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61739 | Gene names | Itga6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA8_HUMAN
|
||||||
| θ value | 6.82597e-32 (rank : 27) | NC score | 0.770471 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53708, Q5VX94 | Gene names | ITGA8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-8 precursor [Contains: Integrin alpha-8 heavy chain; Integrin alpha-8 light chain]. | |||||
|
ITA7_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 28) | NC score | 0.762444 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61738, O88731, O88732, P70350, Q61737, Q61741 | Gene names | Itga7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA6_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 29) | NC score | 0.767880 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA7_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 30) | NC score | 0.762514 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA2B_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 31) | NC score | 0.767370 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08514, O95366, Q14443 | Gene names | ITGA2B, GP2B, ITGAB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA2B_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 32) | NC score | 0.762848 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
MATN1_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 33) | NC score | 0.575979 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
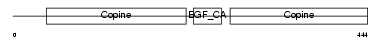
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
COEA1_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 34) | NC score | 0.402060 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
MATN1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 35) | NC score | 0.574616 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 36) | NC score | 0.410676 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 37) | NC score | 0.388423 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 38) | NC score | 0.334612 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
VITRN_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 39) | NC score | 0.577842 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 40) | NC score | 0.390319 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
MATN2_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 41) | NC score | 0.341834 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
VITRN_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 42) | NC score | 0.583728 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
MATN3_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 43) | NC score | 0.405120 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN3_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 44) | NC score | 0.407186 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN4_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 45) | NC score | 0.394718 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 46) | NC score | 0.410308 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
COCH_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 47) | NC score | 0.557242 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 48) | NC score | 0.557629 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 49) | NC score | 0.254817 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 50) | NC score | 0.397204 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 51) | NC score | 0.387322 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
PHLD1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 52) | NC score | 0.481447 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 53) | NC score | 0.493380 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70362 | Gene names | Gpld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
VWF_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 54) | NC score | 0.291658 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 55) | NC score | 0.315262 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CO6A2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 56) | NC score | 0.223160 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
PHLD2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 57) | NC score | 0.462737 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
CO6A1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 58) | NC score | 0.196686 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
CO6A2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 59) | NC score | 0.220346 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 60) | NC score | 0.200346 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.020161 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.016405 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.014833 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.008658 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.027599 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.012889 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.007421 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.016741 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.004184 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
GCSP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.009881 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23378 | Gene names | GLDC, GCSP | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.003269 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
ANGT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.002224 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01019, Q16358, Q16359, Q96F91 | Gene names | AGT, SERPINA8 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.017500 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
GCSP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.008686 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W43 | Gene names | Gldc | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
IKKE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | -0.005818 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14164 | Gene names | IKBKE, IKKE, IKKI, KIAA0151 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase epsilon subunit (EC 2.7.11.10) (I kappa-B kinase epsilon) (IkBKE) (IKK-epsilon) (IKK- E) (Inducible I kappa-B kinase) (IKK-i). | |||||
|
PAPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.005430 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43252, O43841, O75332, Q96FB1, Q96TF4, Q9P1P9, Q9UE98 | Gene names | PAPSS1, ATPSK1, PAPSS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.005429 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60967 | Gene names | Papss1, Asapk, Atpsk1, Papss | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
SETX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.004061 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
ANTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.061156 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.058168 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.066188 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58335, Q59E98, Q5JPE9, Q86UI1, Q8N4J8, Q8NB13, Q96NC7 | Gene names | ANTXR2, CMG2 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 2 precursor (Capillary morphogenesis gene 2 protein) (CMG-2). | |||||
|
CFAB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053019 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051682 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051815 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.053652 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
COGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050944 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
COJA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055808 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CONA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.056669 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
FINC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.057398 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.057202 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050070 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.996836 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAX_MOUSE
|
||||||
| NC score | 0.995859 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QXH4 | Gene names | Itgax | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (CD11c antigen). | |||||
|
ITAD_MOUSE
|
||||||
| NC score | 0.995100 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3V0T4 | Gene names | Itgad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-D precursor (CD11d antigen). | |||||
|
ITAD_HUMAN
|
||||||
| NC score | 0.994244 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13349, Q15575, Q15576 | Gene names | ITGAD | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-D precursor (Leukointegrin alpha D) (ADB2) (CD11d antigen). | |||||
|
ITAX_HUMAN
|
||||||
| NC score | 0.994212 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
ITAL_HUMAN
|
||||||
| NC score | 0.977998 (rank : 7) | θ value | 1.16101e-156 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
ITAL_MOUSE
|
||||||
| NC score | 0.977744 (rank : 8) | θ value | 2.18956e-155 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
ITAE_HUMAN
|
||||||
| NC score | 0.962048 (rank : 9) | θ value | 3.42535e-108 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P38570, Q9NZU9 | Gene names | ITGAE | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Mucosal lymphocyte 1 antigen) (HML-1 antigen) (Integrin alpha-IEL) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITAE_MOUSE
|
||||||
| NC score | 0.961314 (rank : 10) | θ value | 1.48926e-103 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60677 | Gene names | Itgae | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Integrin alpha M290) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA10_HUMAN
|
||||||
| NC score | 0.952443 (rank : 11) | θ value | 1.22112e-97 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
ITA1_HUMAN
|
||||||
| NC score | 0.952221 (rank : 12) | θ value | 4.79078e-102 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P56199 | Gene names | ITGA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-1 (Laminin and collagen receptor) (VLA-1) (CD49a antigen). | |||||
|
ITA11_HUMAN
|
||||||
| NC score | 0.950958 (rank : 13) | θ value | 4.95767e-99 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITA11_MOUSE
|
||||||
| NC score | 0.948418 (rank : 14) | θ value | 8.19079e-94 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P61622 | Gene names | Itga11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITA2_MOUSE
|
||||||
| NC score | 0.947758 (rank : 15) | θ value | 2.08291e-97 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62469, Q62163 | Gene names | Itga2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITA2_HUMAN
|
||||||
| NC score | 0.946551 (rank : 16) | θ value | 1.49272e-95 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17301, Q14595 | Gene names | ITGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITA9_HUMAN
|
||||||
| NC score | 0.845771 (rank : 17) | θ value | 1.91031e-50 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
ITA4_MOUSE
|
||||||
| NC score | 0.843711 (rank : 18) | θ value | 1.66964e-54 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
ITA4_HUMAN
|
||||||
| NC score | 0.839780 (rank : 19) | θ value | 1.01294e-51 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
ITA3_MOUSE
|
||||||
| NC score | 0.805944 (rank : 20) | θ value | 1.12253e-42 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA3_HUMAN
|
||||||
| NC score | 0.805001 (rank : 21) | θ value | 9.83387e-39 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26006 | Gene names | ITGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (FRP-2) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITAV_MOUSE
|
||||||
| NC score | 0.780182 (rank : 22) | θ value | 8.32485e-38 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43406 | Gene names | Itgav | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA5_MOUSE
|
||||||
| NC score | 0.778618 (rank : 23) | θ value | 1.85459e-37 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA5_HUMAN
|
||||||
| NC score | 0.777954 (rank : 24) | θ value | 1.73584e-35 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITAV_HUMAN
|
||||||
| NC score | 0.775737 (rank : 25) | θ value | 1.57e-36 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06756 | Gene names | ITGAV, VNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA6_MOUSE
|
||||||
| NC score | 0.773461 (rank : 26) | θ value | 3.27365e-34 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61739 | Gene names | Itga6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA8_HUMAN
|
||||||
| NC score | 0.770471 (rank : 27) | θ value | 6.82597e-32 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53708, Q5VX94 | Gene names | ITGA8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-8 precursor [Contains: Integrin alpha-8 heavy chain; Integrin alpha-8 light chain]. | |||||
|
ITA6_HUMAN
|
||||||
| NC score | 0.767880 (rank : 28) | θ value | 5.77852e-31 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA2B_HUMAN
|
||||||
| NC score | 0.767370 (rank : 29) | θ value | 9.2256e-29 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08514, O95366, Q14443 | Gene names | ITGA2B, GP2B, ITGAB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA2B_MOUSE
|
||||||
| NC score | 0.762848 (rank : 30) | θ value | 6.61148e-27 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA7_HUMAN
|
||||||
| NC score | 0.762514 (rank : 31) | θ value | 2.42779e-29 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA7_MOUSE
|
||||||
| NC score | 0.762444 (rank : 32) | θ value | 2.59387e-31 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61738, O88731, O88732, P70350, Q61737, Q61741 | Gene names | Itga7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.583728 (rank : 33) | θ value | 6.85773e-16 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.577842 (rank : 34) | θ value | 8.10077e-17 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.575979 (rank : 35) | θ value | 7.82807e-20 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
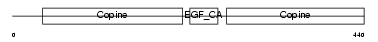
MATN1_HUMAN
|
||||||
| NC score | 0.574616 (rank : 36) | θ value | 5.07402e-19 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
COCH_MOUSE
|
||||||
| NC score | 0.557629 (rank : 37) | θ value | 3.18553e-13 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_HUMAN
|
||||||
| NC score | 0.557242 (rank : 38) | θ value | 1.09485e-13 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
PHLD_MOUSE
|
||||||
| NC score | 0.493380 (rank : 39) | θ value | 9.92553e-07 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70362 | Gene names | Gpld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD1_HUMAN
|
||||||
| NC score | 0.481447 (rank : 40) | θ value | 2.61198e-07 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD2_HUMAN
|
||||||
| NC score | 0.462737 (rank : 41) | θ value | 0.000158464 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.410676 (rank : 42) | θ value | 1.63225e-17 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.410308 (rank : 43) | θ value | 6.41864e-14 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
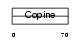
MATN3_HUMAN
|
||||||
| NC score | 0.407186 (rank : 44) | θ value | 5.8054e-15 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
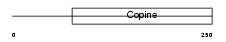
MATN3_MOUSE
|
||||||
| NC score | 0.405120 (rank : 45) | θ value | 2.60593e-15 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.402060 (rank : 46) | θ value | 2.97466e-19 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.397204 (rank : 47) | θ value | 4.30538e-10 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.394718 (rank : 48) | θ value | 9.90251e-15 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.390319 (rank : 49) | θ value | 1.058e-16 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.388423 (rank : 50) | θ value | 3.63628e-17 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.387322 (rank : 51) | θ value | 1.17247e-07 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.341834 (rank : 52) | θ value | 2.35696e-16 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.334612 (rank : 53) | θ value | 6.20254e-17 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.315262 (rank : 54) | θ value | 1.43324e-05 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.291658 (rank : 55) | θ value | 6.43352e-06 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.254817 (rank : 56) | θ value | 8.67504e-11 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CO6A2_MOUSE
|
||||||
| NC score | 0.223160 (rank : 57) | θ value | 2.44474e-05 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A2_HUMAN
|
||||||
| NC score | 0.220346 (rank : 58) | θ value | 0.000786445 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A1_MOUSE
|
||||||
| NC score | 0.200346 (rank : 59) | θ value | 0.0961366 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
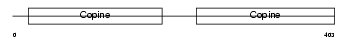
CO6A1_HUMAN
|
||||||
| NC score | 0.196686 (rank : 60) | θ value | 0.000786445 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
ANTR2_HUMAN
|
||||||
| NC score | 0.066188 (rank : 61) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58335, Q59E98, Q5JPE9, Q86UI1, Q8N4J8, Q8NB13, Q96NC7 | Gene names | ANTXR2, CMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 2 precursor (Capillary morphogenesis gene 2 protein) (CMG-2). | |||||
|
ANTR1_HUMAN
|
||||||
| NC score | 0.061156 (rank : 62) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.058168 (rank : 63) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.057398 (rank : 64) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.057202 (rank : 65) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
CONA1_HUMAN
|
||||||
| NC score | 0.056669 (rank : 66) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.055808 (rank : 67) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.053652 (rank : 68) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CFAB_MOUSE
|
||||||
| NC score | 0.053019 (rank : 69) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.051815 (rank : 70) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.051682 (rank : 71) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.050944 (rank : 72) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.050070 (rank : 73) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.027599 (rank : 74) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.020161 (rank : 75) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.017500 (rank : 76) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.016741 (rank : 77) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.016405 (rank : 78) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.014833 (rank : 79) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.012889 (rank : 80) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
GCSP_HUMAN
|
||||||
| NC score | 0.009881 (rank : 81) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23378 | Gene names | GLDC, GCSP | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
GCSP_MOUSE
|
||||||
| NC score | 0.008686 (rank : 82) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W43 | Gene names | Gldc | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.008658 (rank : 83) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.007421 (rank : 84) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
PAPS1_HUMAN
|
||||||
| NC score | 0.005430 (rank : 85) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43252, O43841, O75332, Q96FB1, Q96TF4, Q9P1P9, Q9UE98 | Gene names | PAPSS1, ATPSK1, PAPSS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS1_MOUSE
|
||||||
| NC score | 0.005429 (rank : 86) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60967 | Gene names | Papss1, Asapk, Atpsk1, Papss | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.004184 (rank : 87) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.004061 (rank : 88) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.003269 (rank : 89) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
ANGT_HUMAN
|
||||||
| NC score | 0.002224 (rank : 90) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01019, Q16358, Q16359, Q96F91 | Gene names | AGT, SERPINA8 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensinogen precursor [Contains: Angiotensin-1 (Angiotensin I) (Ang I); Angiotensin-2 (Angiotensin II) (Ang II); Angiotensin-3 (Angiotensin III) (Ang III) (Des-Asp[1]-angiotensin II)]. | |||||
|
IKKE_HUMAN
|
||||||
| NC score | -0.005818 (rank : 91) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14164 | Gene names | IKBKE, IKKE, IKKI, KIAA0151 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase epsilon subunit (EC 2.7.11.10) (I kappa-B kinase epsilon) (IkBKE) (IKK-epsilon) (IKK- E) (Inducible I kappa-B kinase) (IKK-i). | |||||