Please be patient as the page loads
|
DUS8_HUMAN
|
||||||
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DUS8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984860 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 1.4755e-135 (rank : 3) | NC score | 0.970683 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 4) | NC score | 0.925276 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 5) | NC score | 0.926174 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 5.57106e-42 (rank : 6) | NC score | 0.921407 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 7) | NC score | 0.911548 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 8) | NC score | 0.910617 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 9) | NC score | 0.918095 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 10) | NC score | 0.917697 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 11) | NC score | 0.911100 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 12) | NC score | 0.909837 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 13) | NC score | 0.928025 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 5.39604e-37 (rank : 14) | NC score | 0.917600 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 1.20211e-36 (rank : 15) | NC score | 0.917996 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 16) | NC score | 0.907603 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
SSH1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 17) | NC score | 0.762828 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 18) | NC score | 0.854776 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 19) | NC score | 0.855961 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 20) | NC score | 0.745087 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 21) | NC score | 0.747872 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 22) | NC score | 0.777559 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 23) | NC score | 0.859904 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 24) | NC score | 0.832876 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 25) | NC score | 0.860252 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 26) | NC score | 0.760774 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 27) | NC score | 0.851654 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 28) | NC score | 0.832866 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 29) | NC score | 0.773593 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 30) | NC score | 0.852894 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 31) | NC score | 0.856101 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 32) | NC score | 0.833266 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 33) | NC score | 0.850370 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 34) | NC score | 0.840928 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 35) | NC score | 0.778055 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 36) | NC score | 0.819882 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 37) | NC score | 0.818715 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 38) | NC score | 0.761632 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 39) | NC score | 0.776170 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 40) | NC score | 0.771137 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 41) | NC score | 0.071056 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 42) | NC score | 0.767042 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
DUS23_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 43) | NC score | 0.337620 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 44) | NC score | 0.042343 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DUS23_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 45) | NC score | 0.311448 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 46) | NC score | 0.513350 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 47) | NC score | 0.076630 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 48) | NC score | 0.036550 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 49) | NC score | 0.317792 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 50) | NC score | 0.477142 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 51) | NC score | 0.048432 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 52) | NC score | 0.040348 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 53) | NC score | 0.005703 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 54) | NC score | 0.049029 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 55) | NC score | 0.051765 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CC14B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 56) | NC score | 0.280109 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 57) | NC score | 0.021368 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 58) | NC score | 0.033408 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MK15_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 59) | NC score | 0.005731 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
TP4A2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 60) | NC score | 0.129509 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 61) | NC score | 0.129509 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 62) | NC score | 0.044920 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 63) | NC score | 0.028730 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.280196 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 65) | NC score | 0.007988 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 66) | NC score | 0.021314 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.163984 (rank : 67) | NC score | 0.037614 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 68) | NC score | 0.027618 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 69) | NC score | 0.281063 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 70) | NC score | 0.031954 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 71) | NC score | 0.018336 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
TP4A1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.130843 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.130843 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 74) | NC score | 0.027444 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 75) | NC score | 0.028718 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.012961 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
DUS11_MOUSE
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.105361 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.029219 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NUP53_MOUSE
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.033449 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 80) | NC score | 0.157625 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 81) | NC score | 0.153764 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.029872 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.009113 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.255633 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.258645 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.008603 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.029805 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 88) | NC score | 0.018894 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 89) | NC score | 0.017965 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.028315 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.023406 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.011107 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
ZN395_HUMAN
|
||||||
| θ value | 0.813845 (rank : 93) | NC score | 0.032702 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.018373 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.011406 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.007105 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.000089 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.000074 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.028671 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
ICAM4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.027920 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |
|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.002702 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 102) | NC score | 0.016133 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.003559 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.026495 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 105) | NC score | 0.014482 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.017186 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 107) | NC score | 0.018444 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
NU133_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.017945 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUM0, Q9H9W2, Q9NV71, Q9NVC4 | Gene names | NUP133 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup133 (Nucleoporin Nup133) (133 kDa nucleoporin). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.016909 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
5HT7R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | -0.003517 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34969, P78336, P78372, P78516 | Gene names | HTR7 | |||
|
Domain Architecture |
|
|||||
| Description | 5-hydroxytryptamine 7 receptor (5-HT-7) (Serotonin receptor 7) (5-HT- X) (5HT7). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.019268 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 112) | NC score | 0.007421 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 113) | NC score | 0.033123 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 114) | NC score | 0.005657 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 115) | NC score | 0.017185 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.020456 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
RHBD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.010748 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.004308 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.004901 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.026756 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.014667 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.029060 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
LAG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.012704 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61790, Q0VBL2 | Gene names | Lag3 | |||
|
Domain Architecture |
|
|||||
| Description | Lymphocyte activation gene 3 protein precursor (LAG-3) (CD223 antigen). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.001860 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.015571 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.028291 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
K2027_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.011837 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.003287 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.003198 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.017787 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
SYIM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.007648 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.026960 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.028649 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.026211 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.030900 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.010176 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
DCX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.006965 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.010479 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.001280 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.011943 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.025092 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.014093 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
RPC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.009383 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NW08, Q9NW59 | Gene names | POLR3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III subunit 127.6 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 2) (RPC2). | |||||
|
RPC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.009384 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59470, Q6NVF3 | Gene names | Polr3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit 127.6 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 2) (RPC2). | |||||
|
USBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.008742 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.004290 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
KCD18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.006443 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KLF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.002657 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46099, O70261 | Gene names | Klf1, Elkf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 1 (Erythroid krueppel-like transcription factor) (EKLF) (Erythroid transcription factor). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.002432 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.002073 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.019969 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.024935 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.000840 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.008553 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PTCD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.008078 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.016067 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.006115 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.023316 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.014437 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.004549 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
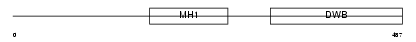
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
ZN414_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004585 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.057070 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.061931 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.064385 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.062964 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.984860 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.970683 (rank : 3) | θ value | 1.4755e-135 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.928025 (rank : 4) | θ value | 1.28434e-38 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.926174 (rank : 5) | θ value | 2.41655e-45 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.925276 (rank : 6) | θ value | 2.41655e-45 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.921407 (rank : 7) | θ value | 5.57106e-42 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.918095 (rank : 8) | θ value | 2.34062e-40 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.917996 (rank : 9) | θ value | 1.20211e-36 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.917697 (rank : 10) | θ value | 5.21438e-40 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.917600 (rank : 11) | θ value | 5.39604e-37 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.911548 (rank : 12) | θ value | 1.62093e-41 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.911100 (rank : 13) | θ value | 8.89434e-40 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.910617 (rank : 14) | θ value | 2.76489e-41 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.909837 (rank : 15) | θ value | 7.52953e-39 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.907603 (rank : 16) | θ value | 3.74554e-30 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.860252 (rank : 17) | θ value | 4.74913e-17 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.859904 (rank : 18) | θ value | 2.7842e-17 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.856101 (rank : 19) | θ value | 2.60593e-15 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.855961 (rank : 20) | θ value | 4.29542e-18 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.854776 (rank : 21) | θ value | 3.28887e-18 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.852894 (rank : 22) | θ value | 5.25075e-16 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.851654 (rank : 23) | θ value | 1.38178e-16 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.850370 (rank : 24) | θ value | 7.58209e-15 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.840928 (rank : 25) | θ value | 2.20605e-14 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.833266 (rank : 26) | θ value | 5.8054e-15 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.832876 (rank : 27) | θ value | 4.74913e-17 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.832866 (rank : 28) | θ value | 1.80466e-16 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.819882 (rank : 29) | θ value | 1.47974e-10 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.818715 (rank : 30) | θ value | 3.29651e-10 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.778055 (rank : 31) | θ value | 1.33837e-11 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.777559 (rank : 32) | θ value | 2.13179e-17 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.776170 (rank : 33) | θ value | 8.11959e-09 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.773593 (rank : 34) | θ value | 3.07829e-16 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.771137 (rank : 35) | θ value | 1.17247e-07 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.767042 (rank : 36) | θ value | 8.40245e-06 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.762828 (rank : 37) | θ value | 5.07402e-19 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.761632 (rank : 38) | θ value | 5.62301e-10 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.760774 (rank : 39) | θ value | 4.74913e-17 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.747872 (rank : 40) | θ value | 1.24977e-17 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.745087 (rank : 41) | θ value | 1.24977e-17 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.513350 (rank : 42) | θ value | 0.00175202 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.477142 (rank : 43) | θ value | 0.00390308 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.337620 (rank : 44) | θ value | 0.00035302 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.317792 (rank : 45) | θ value | 0.00390308 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.311448 (rank : 46) | θ value | 0.00134147 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.281063 (rank : 47) | θ value | 0.21417 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.280196 (rank : 48) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.280109 (rank : 49) | θ value | 0.0563607 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.258645 (rank : 50) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.255633 (rank : 51) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.157625 (rank : 52) | θ value | 0.365318 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.153764 (rank : 53) | θ value | 0.365318 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
TP4A1_HUMAN
|
||||||
| NC score | 0.130843 (rank : 54) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| NC score | 0.130843 (rank : 55) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
TP4A2_HUMAN
|
||||||
| NC score | 0.129509 (rank : 56) | θ value | 0.0961366 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| NC score | 0.129509 (rank : 57) | θ value | 0.0961366 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
DUS11_MOUSE
|
||||||
| NC score | 0.105361 (rank : 58) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.076630 (rank : 59) | θ value | 0.00228821 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.071056 (rank : 60) | θ value | 2.88788e-06 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.064385 (rank : 61) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.062964 (rank : 62) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.061931 (rank : 63) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.057070 (rank : 64) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.051765 (rank : 65) | θ value | 0.0330416 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.049029 (rank : 66) | θ value | 0.0252991 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.048432 (rank : 67) | θ value | 0.00509761 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.044920 (rank : 68) | θ value | 0.125558 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.042343 (rank : 69) | θ value | 0.000461057 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.040348 (rank : 70) | θ value | 0.00869519 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.037614 (rank : 71) | θ value | 0.163984 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.036550 (rank : 72) | θ value | 0.00228821 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NUP53_MOUSE
|
||||||
| NC score | 0.033449 (rank : 73) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.033408 (rank : 74) | θ value | 0.0563607 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.033123 (rank : 75) | θ value | 2.36792 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ZN395_HUMAN
|
||||||
| NC score | 0.032702 (rank : 76) | θ value | 0.813845 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.031954 (rank : 77) | θ value | 0.21417 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.030900 (rank : 78) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.029872 (rank : 79) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.029805 (rank : 80) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.029219 (rank : 81) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.029060 (rank : 82) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.028730 (rank : 83) | θ value | 0.125558 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.028718 (rank : 84) | θ value | 0.279714 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.028671 (rank : 85) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.028649 (rank : 86) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.028315 (rank : 87) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.028291 (rank : 88) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ICAM4_HUMAN
|
||||||
| NC score | 0.027920 (rank : 89) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.027618 (rank : 90) | θ value | 0.21417 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.027444 (rank : 91) | θ value | 0.279714 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.026960 (rank : 92) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.026756 (rank : 93) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.026495 (rank : 94) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.026211 (rank : 95) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.025092 (rank : 96) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.024935 (rank : 97) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.023406 (rank : 98) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.023316 (rank : 99) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.021368 (rank : 100) | θ value | 0.0563607 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.021314 (rank : 101) | θ value | 0.163984 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.020456 (rank : 102) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.019969 (rank : 103) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.019268 (rank : 104) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.018894 (rank : 105) | θ value | 0.62314 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.018444 (rank : 106) | θ value | 1.81305 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.018373 (rank : 107) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.018336 (rank : 108) | θ value | 0.21417 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.017965 (rank : 109) | θ value | 0.62314 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NU133_HUMAN
|
||||||
| NC score | 0.017945 (rank : 110) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUM0, Q9H9W2, Q9NV71, Q9NVC4 | Gene names | NUP133 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup133 (Nucleoporin Nup133) (133 kDa nucleoporin). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.017787 (rank : 111) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.017186 (rank : 112) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.017185 (rank : 113) | θ value | 2.36792 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.016909 (rank : 114) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.016133 (rank : 115) | θ value | 1.38821 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.016067 (rank : 116) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.015571 (rank : 117) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.014667 (rank : 118) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.014482 (rank : 119) | θ value | 1.81305 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.014437 (rank : 120) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.014093 (rank : 121) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.012961 (rank : 122) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
LAG3_MOUSE
|
||||||
| NC score | 0.012704 (rank : 123) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61790, Q0VBL2 | Gene names | Lag3 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte activation gene 3 protein precursor (LAG-3) (CD223 antigen). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.011943 (rank : 124) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
K2027_MOUSE
|
||||||
| NC score | 0.011837 (rank : 125) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.011406 (rank : 126) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.011107 (rank : 127) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
RHBD1_HUMAN
|
||||||
| NC score | 0.010748 (rank : 128) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.010479 (rank : 129) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.010176 (rank : 130) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
RPC2_MOUSE
|
||||||
| NC score | 0.009384 (rank : 131) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59470, Q6NVF3 | Gene names | Polr3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit 127.6 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 2) (RPC2). | |||||
|
RPC2_HUMAN
|
||||||
| NC score | 0.009383 (rank : 132) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NW08, Q9NW59 | Gene names | POLR3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III subunit 127.6 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 2) (RPC2). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.009113 (rank : 133) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
USBP1_HUMAN
|
||||||
| NC score | 0.008742 (rank : 134) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.008603 (rank : 135) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.008553 (rank : 136) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PTCD1_HUMAN
|
||||||
| NC score | 0.008078 (rank : 137) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.007988 (rank : 138) | θ value | 0.163984 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SYIM_HUMAN
|
||||||
| NC score | 0.007648 (rank : 139) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.007421 (rank : 140) | θ value | 2.36792 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.007105 (rank : 141) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.006965 (rank : 142) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.006443 (rank : 143) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.006115 (rank : 144) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.005731 (rank : 145) | θ value | 0.0563607 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.005703 (rank : 146) | θ value | 0.0148317 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.005657 (rank : 147) | θ value | 2.36792 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.004901 (rank : 148) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ZN414_MOUSE
|
||||||
| NC score | 0.004585 (rank : 149) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.004549 (rank : 150) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.004308 (rank : 151) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.004290 (rank : 152) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.003559 (rank : 153) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.003287 (rank : 154) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.003198 (rank : 155) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.002702 (rank : 156) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.002432 (rank : 157) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.002073 (rank : 158) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.001860 (rank : 159) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.001280 (rank : 160) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.000840 (rank : 161) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.000089 (rank : 162) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.000074 (rank : 163) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
KLF1_MOUSE
|
||||||
| NC score | -0.002657 (rank : 164) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46099, O70261 | Gene names | Klf1, Elkf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 1 (Erythroid krueppel-like transcription factor) (EKLF) (Erythroid transcription factor). | |||||
|
5HT7R_HUMAN
|
||||||
| NC score | -0.003517 (rank : 165) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34969, P78336, P78372, P78516 | Gene names | HTR7 | |||
|
Domain Architecture |

|
|||||
| Description | 5-hydroxytryptamine 7 receptor (5-HT-7) (Serotonin receptor 7) (5-HT- X) (5HT7). | |||||