Please be patient as the page loads
|
CC14B_MOUSE
|
||||||
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CC14B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992360 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 1.56915e-153 (rank : 3) | NC score | 0.970202 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 8.0589e-150 (rank : 4) | NC score | 0.973747 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 5) | NC score | 0.610774 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
DUS23_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 6) | NC score | 0.613145 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 7) | NC score | 0.463475 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 8) | NC score | 0.433705 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 9) | NC score | 0.433705 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.445743 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
TP4A2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 11) | NC score | 0.414429 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 12) | NC score | 0.414429 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 13) | NC score | 0.324629 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 14) | NC score | 0.324592 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.333205 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.187457 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 17) | NC score | 0.183924 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.327905 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.301257 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 20) | NC score | 0.295045 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 21) | NC score | 0.295568 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 22) | NC score | 0.317767 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 23) | NC score | 0.314325 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 24) | NC score | 0.295081 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 25) | NC score | 0.297392 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.289247 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 27) | NC score | 0.292003 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.313997 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.303047 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
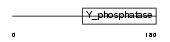
PTEN_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.232394 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
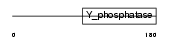
PTEN_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.232395 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.273543 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.295551 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.155141 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.276589 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.280109 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.281833 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS11_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.186619 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 39) | NC score | 0.272852 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
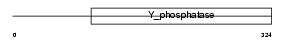
PTN7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 40) | NC score | 0.168004 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.164075 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.139747 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.293879 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.274410 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
MCE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.191815 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60942, O43483, O60257, O60351 | Gene names | RNGTT, CAP1A | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (HCAP1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.155659 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.158712 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.157637 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
MCE1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.177879 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55236 | Gene names | Rngtt, Cap1a | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (MCE1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
PTN7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.164841 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.111732 (rank : 117) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.150125 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.150987 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PTN11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.147194 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.147291 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.119949 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
CSN2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.051160 (rank : 128) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61201, O88950, Q15647, Q6FGP4, Q9BY54, Q9R249, Q9UNI2, Q9UNQ5 | Gene names | COPS2, CSN2, TRIP15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
CSN2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.051160 (rank : 129) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61202, O88950, Q15647, Q3V1W6, Q8R5B0, Q9CWU1, Q9R249, Q9UNQ5 | Gene names | Cops2, Csn2, Trip15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.268554 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.234132 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.118433 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.263982 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.161047 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.160346 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.130100 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.259978 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
ELL3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.031553 (rank : 130) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.115299 (rank : 114) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.166178 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.161959 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.229373 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.248838 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.216743 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.219791 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.116637 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.146068 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.134133 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.256067 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.141823 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.142486 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.133301 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.207923 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.122154 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.126377 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.126036 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.229754 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.013425 (rank : 131) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.147850 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.147202 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.137534 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.110454 (rank : 118) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.111787 (rank : 116) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.124093 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.228018 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.148011 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.133949 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTPRF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.108304 (rank : 119) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009295 (rank : 132) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.133112 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.149621 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.148875 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.114143 (rank : 115) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.146674 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
CDKN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.081798 (rank : 125) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16667, Q99585, Q9BPW7, Q9BY36, Q9C042, Q9C047, Q9C049, Q9C051, Q9C053 | Gene names | CDKN3, CDI1, CIP2, KAP | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 3 (EC 3.1.3.48) (EC 3.1.3.16) (CDK2- associated dual-specificity phosphatase) (Kinase-associated phosphatase) (Cyclin-dependent kinase-interacting protein 2) (Cyclin- dependent kinase interactor 1). | |||||
|
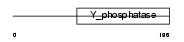
DUS11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.106316 (rank : 121) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75319, Q6AI47, Q9BWE3 | Gene names | DUSP11, PIR1 | |||
|
Domain Architecture |
|
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.253606 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.210844 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.211075 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.059108 (rank : 126) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054710 (rank : 127) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.135159 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.132992 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.129291 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.140388 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.106938 (rank : 120) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.103394 (rank : 122) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.127887 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.103004 (rank : 124) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.103023 (rank : 123) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.123489 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.123087 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.118950 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.119340 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.120478 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.119558 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.122685 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.122686 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
SSH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.171463 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.171802 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.165705 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.166230 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.181294 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.992360 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.973747 (rank : 3) | θ value | 8.0589e-150 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.970202 (rank : 4) | θ value | 1.56915e-153 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.613145 (rank : 5) | θ value | 3.89403e-11 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.610774 (rank : 6) | θ value | 3.89403e-11 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.463475 (rank : 7) | θ value | 4.0297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.445743 (rank : 8) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
TP4A1_HUMAN
|
||||||
| NC score | 0.433705 (rank : 9) | θ value | 5.81887e-07 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| NC score | 0.433705 (rank : 10) | θ value | 5.81887e-07 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
TP4A2_HUMAN
|
||||||
| NC score | 0.414429 (rank : 11) | θ value | 1.09739e-05 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| NC score | 0.414429 (rank : 12) | θ value | 1.09739e-05 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.333205 (rank : 13) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.327905 (rank : 14) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.324629 (rank : 15) | θ value | 2.44474e-05 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.324592 (rank : 16) | θ value | 2.44474e-05 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.317767 (rank : 17) | θ value | 0.00175202 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.314325 (rank : 18) | θ value | 0.00228821 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.313997 (rank : 19) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.303047 (rank : 20) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.301257 (rank : 21) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.297392 (rank : 22) | θ value | 0.00509761 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.295568 (rank : 23) | θ value | 0.00102713 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.295551 (rank : 24) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.295081 (rank : 25) | θ value | 0.00298849 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.295045 (rank : 26) | θ value | 0.00102713 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.293879 (rank : 27) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.292003 (rank : 28) | θ value | 0.00665767 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.289247 (rank : 29) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.281833 (rank : 30) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.280109 (rank : 31) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.276589 (rank : 32) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.274410 (rank : 33) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.273543 (rank : 34) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.272852 (rank : 35) | θ value | 0.0736092 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.268554 (rank : 36) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.263982 (rank : 37) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.259978 (rank : 38) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.256067 (rank : 39) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.253606 (rank : 40) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.248838 (rank : 41) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.234132 (rank : 42) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
PTEN_MOUSE
|
||||||
| NC score | 0.232395 (rank : 43) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_HUMAN
|
||||||
| NC score | 0.232394 (rank : 44) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.229754 (rank : 45) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.229373 (rank : 46) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.228018 (rank : 47) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.219791 (rank : 48) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.216743 (rank : 49) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.211075 (rank : 50) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.210844 (rank : 51) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.207923 (rank : 52) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
MCE1_HUMAN
|
||||||
| NC score | 0.191815 (rank : 53) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60942, O43483, O60257, O60351 | Gene names | RNGTT, CAP1A | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (HCAP1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.187457 (rank : 54) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
DUS11_MOUSE
|
||||||
| NC score | 0.186619 (rank : 55) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.183924 (rank : 56) | θ value | 0.00035302 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.181294 (rank : 57) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
MCE1_MOUSE
|
||||||
| NC score | 0.177879 (rank : 58) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55236 | Gene names | Rngtt, Cap1a | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (MCE1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.171802 (rank : 59) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.171463 (rank : 60) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.168004 (rank : 61) | θ value | 0.0736092 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.166230 (rank : 62) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.166178 (rank : 63) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.165705 (rank : 64) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
PTN7_MOUSE
|
||||||
| NC score | 0.164841 (rank : 65) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.164075 (rank : 66) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.161959 (rank : 67) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.161047 (rank : 68) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.160346 (rank : 69) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.158712 (rank : 70) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.157637 (rank : 71) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.155659 (rank : 72) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.155141 (rank : 73) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.150987 (rank : 74) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.150125 (rank : 75) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
PTN5_HUMAN
|
||||||
| NC score | 0.149621 (rank : 76) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.148875 (rank : 77) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.148011 (rank : 78) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.147850 (rank : 79) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTN11_MOUSE
|
||||||
| NC score | 0.147291 (rank : 80) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.147202 (rank : 81) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTN11_HUMAN
|
||||||
| NC score | 0.147194 (rank : 82) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.146674 (rank : 83) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.146068 (rank : 84) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.142486 (rank : 85) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.141823 (rank : 86) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN1_HUMAN
|
||||||
| NC score | 0.140388 (rank : 87) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.139747 (rank : 88) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.137534 (rank : 89) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.135159 (rank : 90) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.134133 (rank : 91) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTN2_HUMAN
|
||||||
| NC score | 0.133949 (rank : 92) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.133301 (rank : 93) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.133112 (rank : 94) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.132992 (rank : 95) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.130100 (rank : 96) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
PTN18_HUMAN
|
||||||
| NC score | 0.129291 (rank : 97) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.127887 (rank : 98) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.126377 (rank : 99) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.126036 (rank : 100) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.124093 (rank : 101) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.123489 (rank : 102) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| NC score | 0.123087 (rank : 103) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.122686 (rank : 104) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.122685 (rank : 105) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.122154 (rank : 106) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.120478 (rank : 107) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.119949 (rank : 108) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.119558 (rank : 109) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.119340 (rank : 110) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.118950 (rank : 111) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.118433 (rank : 112) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.116637 (rank : 113) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.115299 (rank : 114) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.114143 (rank : 115) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.111787 (rank : 116) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.111732 (rank : 117) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.110454 (rank : 118) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRF_HUMAN
|
||||||
| NC score | 0.108304 (rank : 119) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.106938 (rank : 120) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
DUS11_HUMAN
|
||||||
| NC score | 0.106316 (rank : 121) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75319, Q6AI47, Q9BWE3 | Gene names | DUSP11, PIR1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.103394 (rank : 122) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.103023 (rank : 123) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.103004 (rank : 124) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
CDKN3_HUMAN
|
||||||
| NC score | 0.081798 (rank : 125) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16667, Q99585, Q9BPW7, Q9BY36, Q9C042, Q9C047, Q9C049, Q9C051, Q9C053 | Gene names | CDKN3, CDI1, CIP2, KAP | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 3 (EC 3.1.3.48) (EC 3.1.3.16) (CDK2- associated dual-specificity phosphatase) (Kinase-associated phosphatase) (Cyclin-dependent kinase-interacting protein 2) (Cyclin- dependent kinase interactor 1). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.059108 (rank : 126) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.054710 (rank : 127) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CSN2_HUMAN
|
||||||
| NC score | 0.051160 (rank : 128) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61201, O88950, Q15647, Q6FGP4, Q9BY54, Q9R249, Q9UNI2, Q9UNQ5 | Gene names | COPS2, CSN2, TRIP15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
CSN2_MOUSE
|
||||||
| NC score | 0.051160 (rank : 129) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61202, O88950, Q15647, Q3V1W6, Q8R5B0, Q9CWU1, Q9R249, Q9UNQ5 | Gene names | Cops2, Csn2, Trip15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
ELL3_MOUSE
|
||||||
| NC score | 0.031553 (rank : 130) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.013425 (rank : 131) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.009295 (rank : 132) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||