Please be patient as the page loads
|
SSH2_MOUSE
|
||||||
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SSH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.965279 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.968728 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988966 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 6.65404e-120 (rank : 5) | NC score | 0.956035 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 6.44497e-115 (rank : 6) | NC score | 0.953239 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 7) | NC score | 0.796967 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 8) | NC score | 0.796582 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 9) | NC score | 0.778224 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 10) | NC score | 0.775236 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 11) | NC score | 0.801041 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 12) | NC score | 0.782212 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 13) | NC score | 0.783191 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 14) | NC score | 0.769990 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 15) | NC score | 0.789575 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 16) | NC score | 0.768981 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 17) | NC score | 0.774131 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 18) | NC score | 0.773053 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 19) | NC score | 0.772040 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 20) | NC score | 0.787776 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 21) | NC score | 0.784254 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 22) | NC score | 0.816311 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 23) | NC score | 0.763095 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 24) | NC score | 0.815587 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 25) | NC score | 0.747872 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 26) | NC score | 0.753891 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 27) | NC score | 0.796581 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 28) | NC score | 0.795921 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 29) | NC score | 0.786386 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 30) | NC score | 0.786263 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 31) | NC score | 0.747057 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 32) | NC score | 0.769698 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 33) | NC score | 0.747054 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 34) | NC score | 0.744124 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 35) | NC score | 0.718366 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 36) | NC score | 0.712627 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 37) | NC score | 0.720173 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 38) | NC score | 0.725527 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 39) | NC score | 0.733190 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 40) | NC score | 0.732314 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 41) | NC score | 0.710239 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 42) | NC score | 0.470570 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.439699 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.024770 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 45) | NC score | 0.041726 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.011899 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.006516 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.029860 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.253762 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.033163 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.018229 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TRI55_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.011217 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |
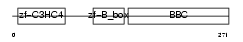
|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
CTCF_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | -0.001618 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
OST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.010471 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.008689 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
HRH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | -0.000427 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |
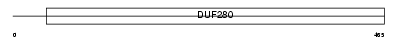
|
|||||
| Description | Histamine H1 receptor. | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.000727 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.003615 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | -0.000087 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GP151_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.009712 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDV0, Q86SN8, Q8NGV2 | Gene names | GPR151, PGR7 | |||
|
Domain Architecture |
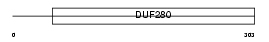
|
|||||
| Description | Probable G-protein coupled receptor 151 (G-protein coupled receptor PGR7) (GPCR-2037). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.000918 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.003256 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.010672 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
WDR47_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.007265 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGF6, Q80TP4 | Gene names | Wdr47, Kiaa0893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 47. | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.192143 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.239278 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
SEPR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.009675 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97321 | Gene names | Fap | |||
|
Domain Architecture |

|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.009846 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.011823 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CASC5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.014178 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
CDT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.017846 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
NCF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.011193 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.182291 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.206655 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.009532 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | -0.000068 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.013311 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
PP2CD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.012130 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
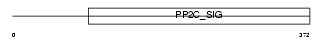
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.014887 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.002470 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CC038_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015005 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JPI3, Q8TC85 | Gene names | C3orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf38. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.008684 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
EPMIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.025120 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
KAD5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.009879 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920P5 | Gene names | Ak5 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.009752 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZFY16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.007376 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.010050 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.004493 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.005711 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.002632 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
DUS23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.199300 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
EPMIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.023276 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
MANS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.013024 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009843 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.008348 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NTKL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.006080 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KG9, Q96G50, Q96KG8, Q96KH1, Q9HAW5, Q9HBL3, Q9NR53 | Gene names | SCYL1, GKLP, NTKL, TAPK, TEIF, TRAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (Teratoma- associated tyrosine kinase) (Telomerase transcriptional element- interacting factor) (Telomerase regulation-associated protein). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.003529 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.002473 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.001481 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
WDR79_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.007148 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BUR4, Q9NW09 | Gene names | WDR79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 79. | |||||
|
CTCF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | -0.002513 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
CX021_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.019218 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf21 homolog. | |||||
|
DUS23_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.178991 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.006178 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.002649 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006379 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006420 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.004443 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBM34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.004197 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5L7 | Gene names | Rbm34, D8Ertd233e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
SPR2G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.020818 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.009765 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.011053 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
FOG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.001589 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
FOG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.001538 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.004932 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.000127 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | -0.000658 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SDCB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.005055 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | -0.003754 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CC14B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.166230 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.050563 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.056807 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.054350 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.988966 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.968728 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.965279 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.956035 (rank : 5) | θ value | 6.65404e-120 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.953239 (rank : 6) | θ value | 6.44497e-115 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.816311 (rank : 7) | θ value | 2.5182e-18 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.815587 (rank : 8) | θ value | 7.32683e-18 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.801041 (rank : 9) | θ value | 3.75424e-22 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.796967 (rank : 10) | θ value | 3.07116e-24 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.796582 (rank : 11) | θ value | 6.84181e-24 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.796581 (rank : 12) | θ value | 6.20254e-17 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.795921 (rank : 13) | θ value | 1.058e-16 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.789575 (rank : 14) | θ value | 1.5773e-20 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.787776 (rank : 15) | θ value | 5.07402e-19 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.786386 (rank : 16) | θ value | 6.41864e-14 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.786263 (rank : 17) | θ value | 8.38298e-14 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.784254 (rank : 18) | θ value | 2.5182e-18 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.783191 (rank : 19) | θ value | 7.0802e-21 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.782212 (rank : 20) | θ value | 7.0802e-21 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.778224 (rank : 21) | θ value | 3.39556e-23 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.775236 (rank : 22) | θ value | 3.39556e-23 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.774131 (rank : 23) | θ value | 5.99374e-20 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.773053 (rank : 24) | θ value | 1.33526e-19 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.772040 (rank : 25) | θ value | 1.33526e-19 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.769990 (rank : 26) | θ value | 9.24701e-21 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.769698 (rank : 27) | θ value | 2.28291e-11 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.768981 (rank : 28) | θ value | 2.69047e-20 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.763095 (rank : 29) | θ value | 4.29542e-18 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.753891 (rank : 30) | θ value | 1.24977e-17 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.747872 (rank : 31) | θ value | 1.24977e-17 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.747057 (rank : 32) | θ value | 1.74796e-11 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.747054 (rank : 33) | θ value | 3.89403e-11 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.744124 (rank : 34) | θ value | 1.133e-10 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.733190 (rank : 35) | θ value | 1.06045e-08 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.732314 (rank : 36) | θ value | 5.26297e-08 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.725527 (rank : 37) | θ value | 8.11959e-09 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.720173 (rank : 38) | θ value | 3.64472e-09 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.718366 (rank : 39) | θ value | 4.30538e-10 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.712627 (rank : 40) | θ value | 1.25267e-09 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.710239 (rank : 41) | θ value | 4.45536e-07 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.470570 (rank : 42) | θ value | 0.00020696 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.439699 (rank : 43) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.253762 (rank : 44) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.239278 (rank : 45) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.206655 (rank : 46) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.199300 (rank : 47) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.192143 (rank : 48) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.182291 (rank : 49) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.178991 (rank : 50) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.166230 (rank : 51) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.056807 (rank : 52) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.054350 (rank : 53) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
DUS11_MOUSE
|
||||||
| NC score | 0.050563 (rank : 54) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.041726 (rank : 55) | θ value | 0.0431538 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.033163 (rank : 56) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.029860 (rank : 57) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
EPMIP_MOUSE
|
||||||
| NC score | 0.025120 (rank : 58) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.024770 (rank : 59) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
EPMIP_HUMAN
|
||||||
| NC score | 0.023276 (rank : 60) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
SPR2G_HUMAN
|
||||||
| NC score | 0.020818 (rank : 61) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
CX021_MOUSE
|
||||||
| NC score | 0.019218 (rank : 62) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf21 homolog. | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.018229 (rank : 63) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
CDT1_MOUSE
|
||||||
| NC score | 0.017846 (rank : 64) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
CC038_HUMAN
|
||||||
| NC score | 0.015005 (rank : 65) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JPI3, Q8TC85 | Gene names | C3orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf38. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.014887 (rank : 66) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.014178 (rank : 67) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.013311 (rank : 68) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MANS1_HUMAN
|
||||||
| NC score | 0.013024 (rank : 69) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.012130 (rank : 70) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.011899 (rank : 71) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.011823 (rank : 72) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TRI55_HUMAN
|
||||||
| NC score | 0.011217 (rank : 73) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
NCF2_HUMAN
|
||||||
| NC score | 0.011193 (rank : 74) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.011053 (rank : 75) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.010672 (rank : 76) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
OST4_HUMAN
|
||||||
| NC score | 0.010471 (rank : 77) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.010050 (rank : 78) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
KAD5_MOUSE
|
||||||
| NC score | 0.009879 (rank : 79) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920P5 | Gene names | Ak5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.009846 (rank : 80) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.009843 (rank : 81) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.009765 (rank : 82) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.009752 (rank : 83) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
GP151_HUMAN
|
||||||
| NC score | 0.009712 (rank : 84) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDV0, Q86SN8, Q8NGV2 | Gene names | GPR151, PGR7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 151 (G-protein coupled receptor PGR7) (GPCR-2037). | |||||
|
SEPR_MOUSE
|
||||||
| NC score | 0.009675 (rank : 85) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97321 | Gene names | Fap | |||
|
Domain Architecture |

|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.009532 (rank : 86) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.008689 (rank : 87) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.008684 (rank : 88) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.008348 (rank : 89) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ZFY16_MOUSE
|
||||||
| NC score | 0.007376 (rank : 90) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
WDR47_MOUSE
|
||||||
| NC score | 0.007265 (rank : 91) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGF6, Q80TP4 | Gene names | Wdr47, Kiaa0893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 47. | |||||
|
WDR79_HUMAN
|
||||||
| NC score | 0.007148 (rank : 92) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BUR4, Q9NW09 | Gene names | WDR79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 79. | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.006516 (rank : 93) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.006420 (rank : 94) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.006379 (rank : 95) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.006178 (rank : 96) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
NTKL_HUMAN
|
||||||
| NC score | 0.006080 (rank : 97) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KG9, Q96G50, Q96KG8, Q96KH1, Q9HAW5, Q9HBL3, Q9NR53 | Gene names | SCYL1, GKLP, NTKL, TAPK, TEIF, TRAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like protein (SCY1-like protein 1) (Teratoma- associated tyrosine kinase) (Telomerase transcriptional element- interacting factor) (Telomerase regulation-associated protein). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.005711 (rank : 98) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SDCB1_MOUSE
|
||||||
| NC score | 0.005055 (rank : 99) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.004932 (rank : 100) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.004493 (rank : 101) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.004443 (rank : 102) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBM34_MOUSE
|
||||||
| NC score | 0.004197 (rank : 103) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5L7 | Gene names | Rbm34, D8Ertd233e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.003615 (rank : 104) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.003529 (rank : 105) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.003256 (rank : 106) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.002649 (rank : 107) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.002632 (rank : 108) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.002473 (rank : 109) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.002470 (rank : 110) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
FOG2_HUMAN
|
||||||
| NC score | 0.001589 (rank : 111) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
FOG2_MOUSE
|
||||||
| NC score | 0.001538 (rank : 112) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.001481 (rank : 113) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.000918 (rank : 114) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.000727 (rank : 115) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.000127 (rank : 116) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | -0.000068 (rank : 117) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | -0.000087 (rank : 118) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
HRH1_HUMAN
|
||||||
| NC score | -0.000427 (rank : 119) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |

|
|||||
| Description | Histamine H1 receptor. | |||||
|
RAI14_MOUSE
|
||||||
| NC score | -0.000658 (rank : 120) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CTCF_MOUSE
|
||||||
| NC score | -0.001618 (rank : 121) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
CTCF_HUMAN
|
||||||
| NC score | -0.002513 (rank : 122) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | -0.003754 (rank : 123) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||