Please be patient as the page loads
|
T22D4_MOUSE
|
||||||
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
T22D4_MOUSE
|
||||||
| θ value | 3.85598e-160 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 6.59259e-152 (rank : 2) | NC score | 0.847733 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
T22D1_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 3) | NC score | 0.823703 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
T22D1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 4) | NC score | 0.821950 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15714, O00666, Q96JS5 | Gene names | TSC22D1, TGFB1I4, TSC22 | |||
|
Domain Architecture |
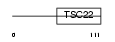
|
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog) (Cerebral protein 2). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 5) | NC score | 0.653823 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
T22D3_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 6) | NC score | 0.812246 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |
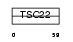
|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
T22D3_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 7) | NC score | 0.808116 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z2S7, Q3UNI6, Q8K160, Q9EQN0, Q9EQN1, Q9EQN2 | Gene names | Tsc22d3, Dsip1, Dsipi, Gilz | |||
|
Domain Architecture |
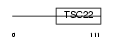
|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (TSC22-related-inducible leucine zipper 3) (Tilz3). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 8) | NC score | 0.111582 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 9) | NC score | 0.115593 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.082254 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.054822 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.035761 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.049642 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.120238 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.120290 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.033227 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.032773 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.016800 (rank : 116) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
ST18_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.043751 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.089481 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.088443 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.042220 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.022774 (rank : 109) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.103625 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.097118 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.100715 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.017877 (rank : 113) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
IPF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.015144 (rank : 118) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52946 | Gene names | Ipf1, Pdx1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin promoter factor 1 (IPF-1) (Islet/duodenum homeobox 1) (IDX-1) (Somatostatin-transactivating factor 1) (STF-1) (Pancreas/duodenum homeobox 1). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.058290 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.071902 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TFG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.068436 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.032253 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.070237 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.065899 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.041746 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.011509 (rank : 126) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.035142 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.024512 (rank : 106) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.058160 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.091184 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ST18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.037181 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.092953 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DCMC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.066991 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J39, Q7TNL6 | Gene names | Mlycd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl-CoA decarboxylase, mitochondrial precursor (EC 4.1.1.9) (MCD). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.059619 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.093311 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.069300 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.047274 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.023949 (rank : 107) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.038810 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.017720 (rank : 114) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.037550 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.060217 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.059750 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.008580 (rank : 130) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.022011 (rank : 111) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.028924 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.038907 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MYCBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.124560 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99417, Q96HE2 | Gene names | MYCBP, AMY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
TBX21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.011456 (rank : 127) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.052027 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.048618 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.008228 (rank : 132) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.026960 (rank : 104) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.013136 (rank : 123) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.035769 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
NID2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.005841 (rank : 133) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.043650 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
DEPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.022014 (rank : 110) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2F3, Q8BU66 | Gene names | Depp, Fseg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEPP (Fat-specific expressed gene protein). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.023133 (rank : 108) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.030596 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
M3K12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011786 (rank : 125) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.030687 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.016175 (rank : 117) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.017681 (rank : 115) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.003091 (rank : 134) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.058297 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.058424 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.014157 (rank : 120) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COE4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.008432 (rank : 131) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQW3, Q5JY53, Q9NUB6, Q9P2A6 | Gene names | EBF4, COE4, KIAA1442 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.038307 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.038362 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.013468 (rank : 122) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.011174 (rank : 128) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
K0056_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.012685 (rank : 124) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
KIRR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.010343 (rank : 129) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
KLF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | -0.000166 (rank : 135) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46099, O70261 | Gene names | Klf1, Elkf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 1 (Erythroid krueppel-like transcription factor) (EKLF) (Erythroid transcription factor). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.014058 (rank : 121) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.020226 (rank : 112) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RTN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.026764 (rank : 105) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |
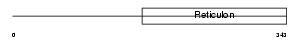
|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.030260 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.014674 (rank : 119) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | -0.000995 (rank : 136) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.102764 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.070031 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.054098 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.055720 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CITE4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.056229 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051185 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.066240 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050500 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.052169 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.051268 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052247 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.058225 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050531 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.061986 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070580 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.062011 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057733 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.090266 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.056952 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055829 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.054306 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.060662 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.083107 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.090928 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.071934 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.080294 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.061660 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.080063 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.068864 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.066117 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.063594 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053132 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.050081 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052346 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.072064 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.059417 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
WASF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060030 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.076075 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.070503 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052327 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.066716 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.057507 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.067829 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.076679 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.85598e-160 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.847733 (rank : 2) | θ value | 6.59259e-152 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
T22D1_MOUSE
|
||||||
| NC score | 0.823703 (rank : 3) | θ value | 9.87957e-23 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
T22D1_HUMAN
|
||||||
| NC score | 0.821950 (rank : 4) | θ value | 1.6852e-22 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15714, O00666, Q96JS5 | Gene names | TSC22D1, TGFB1I4, TSC22 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog) (Cerebral protein 2). | |||||
|
T22D3_HUMAN
|
||||||
| NC score | 0.812246 (rank : 5) | θ value | 3.17815e-21 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
T22D3_MOUSE
|
||||||
| NC score | 0.808116 (rank : 6) | θ value | 2.69047e-20 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z2S7, Q3UNI6, Q8K160, Q9EQN0, Q9EQN1, Q9EQN2 | Gene names | Tsc22d3, Dsip1, Dsipi, Gilz | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (TSC22-related-inducible leucine zipper 3) (Tilz3). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.653823 (rank : 7) | θ value | 2.43343e-21 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
MYCBP_HUMAN
|
||||||
| NC score | 0.124560 (rank : 8) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99417, Q96HE2 | Gene names | MYCBP, AMY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.120290 (rank : 9) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.120238 (rank : 10) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.115593 (rank : 11) | θ value | 0.00134147 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.111582 (rank : 12) | θ value | 0.000786445 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.103625 (rank : 13) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.102764 (rank : 14) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.100715 (rank : 15) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.097118 (rank : 16) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.093311 (rank : 17) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.092953 (rank : 18) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.091184 (rank : 19) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.090928 (rank : 20) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.090266 (rank : 21) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.089481 (rank : 22) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.088443 (rank : 23) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.083107 (rank : 24) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.082254 (rank : 25) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.080294 (rank : 26) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.080063 (rank : 27) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.076679 (rank : 28) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.076075 (rank : 29) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.072064 (rank : 30) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.071934 (rank : 31) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.071902 (rank : 32) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.070580 (rank : 33) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.070503 (rank : 34) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.070237 (rank : 35) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.070031 (rank : 36) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.069300 (rank : 37) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.068864 (rank : 38) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TFG_HUMAN
|
||||||
| NC score | 0.068436 (rank : 39) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.067829 (rank : 40) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
DCMC_MOUSE
|
||||||
| NC score | 0.066991 (rank : 41) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J39, Q7TNL6 | Gene names | Mlycd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malonyl-CoA decarboxylase, mitochondrial precursor (EC 4.1.1.9) (MCD). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.066716 (rank : 42) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.066240 (rank : 43) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.066117 (rank : 44) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.065899 (rank : 45) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.063594 (rank : 46) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.062011 (rank : 47) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.061986 (rank : 48) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.061660 (rank : 49) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.060662 (rank : 50) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.060217 (rank : 51) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WASF1_MOUSE
|
||||||
| NC score | 0.060030 (rank : 52) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.059750 (rank : 53) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.059619 (rank : 54) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
WASF1_HUMAN
|
||||||
| NC score | 0.059417 (rank : 55) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.058424 (rank : 56) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.058297 (rank : 57) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.058290 (rank : 58) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.058225 (rank : 59) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.058160 (rank : 60) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.057733 (rank : 61) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.057507 (rank : 62) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056952 (rank : 63) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CITE4_MOUSE
|
||||||
| NC score | 0.056229 (rank : 64) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.055829 (rank : 65) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.055720 (rank : 66) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.054822 (rank : 67) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.054306 (rank : 68) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.054098 (rank : 69) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.053132 (rank : 70) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.052346 (rank : 71) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.052327 (rank : 72) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.052247 (rank : 73) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.052169 (rank : 74) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.052027 (rank : 75) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.051268 (rank : 76) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.051185 (rank : 77) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.050531 (rank : 78) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.050500 (rank : 79) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.050081 (rank : 80) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.049642 (rank : 81) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.048618 (rank : 82) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.047274 (rank : 83) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
ST18_MOUSE
|
||||||
| NC score | 0.043751 (rank : 84) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.043650 (rank : 85) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.042220 (rank : 86) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.041746 (rank : 87) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.038907 (rank : 88) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.038810 (rank : 89) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.038362 (rank : 90) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.038307 (rank : 91) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.037550 (rank : 92) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.037181 (rank : 93) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.035769 (rank : 94) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.035761 (rank : 95) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.035142 (rank : 96) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.033227 (rank : 97) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.032773 (rank : 98) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.032253 (rank : 99) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.030687 (rank : 100) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.030596 (rank : 101) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.030260 (rank : 102) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.028924 (rank : 103) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.026960 (rank : 104) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
RTN2_HUMAN
|
||||||
| NC score | 0.026764 (rank : 105) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.024512 (rank : 106) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.023949 (rank : 107) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.023133 (rank : 108) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.022774 (rank : 109) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
DEPP_MOUSE
|
||||||
| NC score | 0.022014 (rank : 110) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2F3, Q8BU66 | Gene names | Depp, Fseg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEPP (Fat-specific expressed gene protein). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.022011 (rank : 111) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.020226 (rank : 112) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.017877 (rank : 113) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.017720 (rank : 114) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.017681 (rank : 115) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.016800 (rank : 116) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.016175 (rank : 117) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
IPF1_MOUSE
|
||||||
| NC score | 0.015144 (rank : 118) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52946 | Gene names | Ipf1, Pdx1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin promoter factor 1 (IPF-1) (Islet/duodenum homeobox 1) (IDX-1) (Somatostatin-transactivating factor 1) (STF-1) (Pancreas/duodenum homeobox 1). | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.014674 (rank : 119) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.014157 (rank : 120) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.014058 (rank : 121) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.013468 (rank : 122) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.013136 (rank : 123) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.012685 (rank : 124) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
M3K12_HUMAN
|
||||||
| NC score | 0.011786 (rank : 125) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.011509 (rank : 126) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
TBX21_MOUSE
|
||||||
| NC score | 0.011456 (rank : 127) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.011174 (rank : 128) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
KIRR1_HUMAN
|
||||||
| NC score | 0.010343 (rank : 129) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.008580 (rank : 130) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
COE4_HUMAN
|
||||||
| NC score | 0.008432 (rank : 131) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQW3, Q5JY53, Q9NUB6, Q9P2A6 | Gene names | EBF4, COE4, KIAA1442 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.008228 (rank : 132) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.005841 (rank : 133) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.003091 (rank : 134) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
KLF1_MOUSE
|
||||||
| NC score | -0.000166 (rank : 135) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46099, O70261 | Gene names | Klf1, Elkf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 1 (Erythroid krueppel-like transcription factor) (EKLF) (Erythroid transcription factor). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000995 (rank : 136) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||