Please be patient as the page loads
|
T22D1_MOUSE
|
||||||
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
T22D1_MOUSE
|
||||||
| θ value | 1.78386e-56 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
T22D1_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 2) | NC score | 0.997575 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15714, O00666, Q96JS5 | Gene names | TSC22D1, TGFB1I4, TSC22 | |||
|
Domain Architecture |
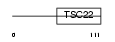
|
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog) (Cerebral protein 2). | |||||
|
T22D3_MOUSE
|
||||||
| θ value | 1.08726e-37 (rank : 3) | NC score | 0.977639 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2S7, Q3UNI6, Q8K160, Q9EQN0, Q9EQN1, Q9EQN2 | Gene names | Tsc22d3, Dsip1, Dsipi, Gilz | |||
|
Domain Architecture |
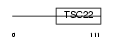
|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (TSC22-related-inducible leucine zipper 3) (Tilz3). | |||||
|
T22D3_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 4) | NC score | 0.977919 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |
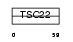
|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 5) | NC score | 0.714998 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 6) | NC score | 0.712738 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 7) | NC score | 0.823703 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.062647 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.062740 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.041360 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
MYCBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.157639 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99417, Q96HE2 | Gene names | MYCBP, AMY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.013233 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
AP3M1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.030711 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.030732 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
PCDG6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.005418 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||
|
TAK1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.050019 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57077 | Gene names | TAK1L, C21orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
NGLY1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.034039 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IV0, Q59FB1, Q6PJD8, Q9BVR8, Q9NR70 | Gene names | NGLY1, PNG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (hPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.025552 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.028379 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
K22O_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.005755 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.015340 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.055888 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.023185 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.023121 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.022938 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.022967 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.018694 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.050850 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.014479 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.012343 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.012179 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.017608 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.014711 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.005930 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
TAK1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.032404 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58500 | Gene names | Tak1l, ORF63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
MYCBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.050612 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQS3, O88308 | Gene names | Mycbp, Amy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
T22D1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.78386e-56 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
T22D1_HUMAN
|
||||||
| NC score | 0.997575 (rank : 2) | θ value | 4.39379e-55 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15714, O00666, Q96JS5 | Gene names | TSC22D1, TGFB1I4, TSC22 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog) (Cerebral protein 2). | |||||
|
T22D3_HUMAN
|
||||||
| NC score | 0.977919 (rank : 3) | θ value | 1.85459e-37 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
T22D3_MOUSE
|
||||||
| NC score | 0.977639 (rank : 4) | θ value | 1.08726e-37 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2S7, Q3UNI6, Q8K160, Q9EQN0, Q9EQN1, Q9EQN2 | Gene names | Tsc22d3, Dsip1, Dsipi, Gilz | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (TSC22-related-inducible leucine zipper 3) (Tilz3). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.823703 (rank : 5) | θ value | 9.87957e-23 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.714998 (rank : 6) | θ value | 6.84181e-24 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.712738 (rank : 7) | θ value | 4.43474e-23 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
MYCBP_HUMAN
|
||||||
| NC score | 0.157639 (rank : 8) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99417, Q96HE2 | Gene names | MYCBP, AMY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.062740 (rank : 9) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.062647 (rank : 10) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.055888 (rank : 11) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.050850 (rank : 12) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
MYCBP_MOUSE
|
||||||
| NC score | 0.050612 (rank : 13) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQS3, O88308 | Gene names | Mycbp, Amy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
TAK1L_HUMAN
|
||||||
| NC score | 0.050019 (rank : 14) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57077 | Gene names | TAK1L, C21orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.041360 (rank : 15) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NGLY1_HUMAN
|
||||||
| NC score | 0.034039 (rank : 16) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IV0, Q59FB1, Q6PJD8, Q9BVR8, Q9NR70 | Gene names | NGLY1, PNG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (hPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
TAK1L_MOUSE
|
||||||
| NC score | 0.032404 (rank : 17) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58500 | Gene names | Tak1l, ORF63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TAK1-like protein. | |||||
|
AP3M1_MOUSE
|
||||||
| NC score | 0.030732 (rank : 18) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_HUMAN
|
||||||
| NC score | 0.030711 (rank : 19) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.028379 (rank : 20) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.025552 (rank : 21) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.023185 (rank : 22) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.023121 (rank : 23) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.022967 (rank : 24) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.022938 (rank : 25) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.018694 (rank : 26) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.017608 (rank : 27) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.015340 (rank : 28) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.014711 (rank : 29) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.014479 (rank : 30) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.013233 (rank : 31) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.012343 (rank : 32) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.012179 (rank : 33) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.005930 (rank : 34) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
K22O_HUMAN
|
||||||
| NC score | 0.005755 (rank : 35) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
PCDG6_HUMAN
|
||||||
| NC score | 0.005418 (rank : 36) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||