Please be patient as the page loads
|
ARFG3_MOUSE
|
||||||
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARFG3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993833 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
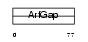
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 3) | NC score | 0.844599 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
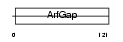
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG1_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 4) | NC score | 0.832707 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
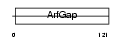
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 5) | NC score | 0.775731 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 6) | NC score | 0.777092 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 7) | NC score | 0.774979 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 8) | NC score | 0.774611 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 9) | NC score | 0.574080 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 10) | NC score | 0.561761 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 11) | NC score | 0.609028 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 12) | NC score | 0.613953 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 13) | NC score | 0.597540 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 14) | NC score | 0.605397 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 15) | NC score | 0.583623 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 16) | NC score | 0.605175 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 17) | NC score | 0.579360 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 18) | NC score | 0.572804 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 19) | NC score | 0.517091 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 20) | NC score | 0.570076 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 21) | NC score | 0.567071 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 22) | NC score | 0.520161 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 23) | NC score | 0.714477 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 24) | NC score | 0.587294 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 25) | NC score | 0.615977 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
GIT2_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 26) | NC score | 0.548953 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |
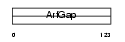
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 27) | NC score | 0.536609 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 28) | NC score | 0.544517 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 29) | NC score | 0.534963 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 30) | NC score | 0.520514 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 31) | NC score | 0.531283 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 32) | NC score | 0.530237 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
CENA2_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 33) | NC score | 0.675433 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENA2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 34) | NC score | 0.665187 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 35) | NC score | 0.522131 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
NUPL_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 36) | NC score | 0.526127 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.045051 (rank : 159) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.032333 (rank : 161) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.412114 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.392406 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
CD3E_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.052163 (rank : 136) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22646 | Gene names | Cd3e | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD3 epsilon chain precursor (T-cell surface antigen T3/Leu-4 epsilon chain). | |||||
|
ALKB5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.031316 (rank : 163) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P6C2, Q9NXD6 | Gene names | ALKBH5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkylated repair protein alkB homolog 5. | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.018487 (rank : 169) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.013396 (rank : 182) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.032253 (rank : 162) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.050544 (rank : 152) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.017990 (rank : 171) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.017390 (rank : 173) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.021564 (rank : 166) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.025425 (rank : 164) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
INSM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.004723 (rank : 192) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.012286 (rank : 184) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PRDM8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.017570 (rank : 172) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ97 | Gene names | Prdm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.013751 (rank : 181) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.024309 (rank : 165) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.039496 (rank : 160) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.014673 (rank : 178) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.018451 (rank : 170) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.009546 (rank : 187) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CLC6A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.011307 (rank : 185) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.016984 (rank : 175) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.013376 (rank : 183) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
FOXC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.003552 (rank : 194) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61850, P97948, Q63869, Q8C694 | Gene names | Foxc2, Fkh14, Fkhl14, Mfh1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
IRF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.010297 (rank : 186) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.014916 (rank : 177) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.019654 (rank : 167) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | -0.000433 (rank : 196) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.018909 (rank : 168) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | -0.001156 (rank : 197) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.008451 (rank : 188) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.002474 (rank : 195) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
ZN524_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | -0.003131 (rank : 198) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96C55, Q6NW31, Q96IL7 | Gene names | ZNF524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 524. | |||||
|
CCDC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.014673 (rank : 179) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.017245 (rank : 174) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.007488 (rank : 189) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CSTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.003564 (rank : 193) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
ERF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.005551 (rank : 191) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005680 (rank : 190) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TS101_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.016890 (rank : 176) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.014188 (rank : 180) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ACBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055620 (rank : 109) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ACBD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.054450 (rank : 116) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
AN13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051892 (rank : 141) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YJ7, Q8N7S9 | Gene names | ANKRD13B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
AN13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051969 (rank : 138) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5F259, Q6IR40 | Gene names | Ankrd13b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
AN36A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051944 (rank : 140) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.059339 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.059360 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050306 (rank : 155) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050405 (rank : 153) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.058743 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.059739 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.063811 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
ANKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.058770 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANKR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053797 (rank : 122) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NU02, Q9H6Y9 | Gene names | ANKRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANKR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.053415 (rank : 126) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.061543 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050385 (rank : 154) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
ANR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.062822 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059999 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051553 (rank : 144) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.056280 (rank : 106) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ANR23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058109 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055576 (rank : 110) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052804 (rank : 132) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053400 (rank : 127) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.060233 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.058953 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ANR37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.071320 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z713 | Gene names | ANKRD37, LPR2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37 (Low-density lipoprotein receptor-related protein 2-binding protein) (hLrp2bp). | |||||
|
ANR37_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.082316 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569N2, Q8CHS0 | Gene names | Ankrd37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37. | |||||
|
ANR39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.075641 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q53RE8, Q59FU2, Q8N5X5, Q9P0S5 | Gene names | ANKRD39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.076881 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D2X0 | Gene names | Ankrd39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.086928 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.059840 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055739 (rank : 108) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.055198 (rank : 112) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.084635 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.083718 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTZ5, Q1LZK9, Q8BV97 | Gene names | Ankrd46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.053927 (rank : 120) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
ANR49_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051127 (rank : 147) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054595 (rank : 114) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058513 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.058599 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.058981 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
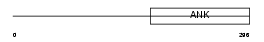
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.059843 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANS4B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.066524 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ANS4B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.072978 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ASB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.061633 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |
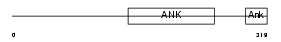
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058767 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.051806 (rank : 143) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXK4, Q52LK4, Q6ISF9 | Gene names | ASB12 | |||
|
Domain Architecture |
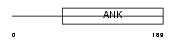
|
|||||
| Description | Ankyrin repeat and SOCS box protein 12 (ASB-12). | |||||
|
ASB12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052307 (rank : 134) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D738 | Gene names | Asb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 12 (ASB-12). | |||||
|
ASB13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.050111 (rank : 157) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VBX0 | Gene names | Asb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
ASB16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.053094 (rank : 128) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |
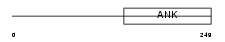
|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051862 (rank : 142) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.052229 (rank : 135) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053804 (rank : 121) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053000 (rank : 130) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |
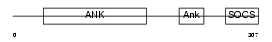
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.053001 (rank : 129) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |
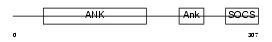
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.057900 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |
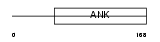
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.057449 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |
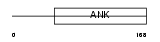
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056206 (rank : 107) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.053485 (rank : 125) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
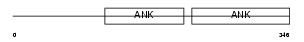
BCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.055218 (rank : 111) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.056341 (rank : 105) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
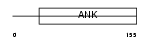
CDN2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.065611 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
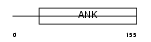
CDN2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.056406 (rank : 104) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.052429 (rank : 133) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.051965 (rank : 139) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.054285 (rank : 117) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.053644 (rank : 123) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CT086_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.104613 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050586 (rank : 151) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.099813 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.099324 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.088011 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.096975 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.054183 (rank : 118) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050604 (rank : 150) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
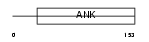
GABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.072256 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.071699 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |
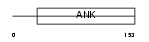
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
GABP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.073293 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P81069 | Gene names | Gabpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GA-binding protein beta-2-1 chain (GABP subunit beta-2-1) (GABPB2-1). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050688 (rank : 149) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.066227 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.066998 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
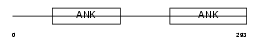
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.077296 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
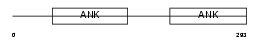
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
MTPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.066869 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58546 | Gene names | MTPN | |||
|
Domain Architecture |
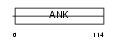
|
|||||
| Description | Myotrophin (Protein V-1). | |||||
|
MTPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068730 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62774, P80144, Q543M6, Q9DCN8 | Gene names | Mtpn, Gcdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotrophin (Protein V-1) (Granule cell differentiation protein). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.060673 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.061298 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055114 (rank : 113) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.102542 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.112508 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.056748 (rank : 103) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052805 (rank : 131) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.085847 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.083862 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.058852 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.065207 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.066185 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.067961 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
POTE8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050190 (rank : 156) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6S8J7, Q6S8J6 | Gene names | POTE8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 8. | |||||
|
PP16A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.053557 (rank : 124) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |
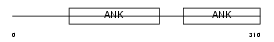
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
PP16B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.050823 (rank : 148) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHQ3 | Gene names | Ppp1r16b | |||
|
Domain Architecture |
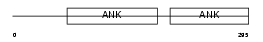
|
|||||
| Description | Protein phosphatase 1 regulatory inhibitor subunit 16B (TGF-beta- inhibited membrane-associated protein) (CAAX box protein TIMAP). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.059782 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
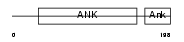
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.058779 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
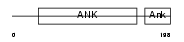
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.059756 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.057046 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.054571 (rank : 115) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.054127 (rank : 119) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.050019 (rank : 158) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.051392 (rank : 145) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.058340 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.096860 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.052073 (rank : 137) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.051346 (rank : 146) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.063688 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.060581 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
USH1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.061185 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
USH1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 198) | NC score | 0.061455 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.993833 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.844599 (rank : 3) | θ value | 1.47289e-26 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.832707 (rank : 4) | θ value | 3.62785e-25 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.777092 (rank : 5) | θ value | 3.40345e-15 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.775731 (rank : 6) | θ value | 3.40345e-15 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.774979 (rank : 7) | θ value | 3.40345e-15 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.774611 (rank : 8) | θ value | 4.44505e-15 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.714477 (rank : 9) | θ value | 9.59137e-10 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_MOUSE
|
||||||
| NC score | 0.675433 (rank : 10) | θ value | 1.99992e-07 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.665187 (rank : 11) | θ value | 2.21117e-06 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.615977 (rank : 12) | θ value | 2.13673e-09 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.613953 (rank : 13) | θ value | 8.38298e-14 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.609028 (rank : 14) | θ value | 8.38298e-14 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.605397 (rank : 15) | θ value | 1.42992e-13 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.605175 (rank : 16) | θ value | 1.86753e-13 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.597540 (rank : 17) | θ value | 1.09485e-13 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.587294 (rank : 18) | θ value | 1.25267e-09 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.583623 (rank : 19) | θ value | 1.86753e-13 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.579360 (rank : 20) | θ value | 4.16044e-13 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.574080 (rank : 21) | θ value | 9.90251e-15 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.572804 (rank : 22) | θ value | 2.98157e-11 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.570076 (rank : 23) | θ value | 4.30538e-10 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.567071 (rank : 24) | θ value | 4.30538e-10 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.561761 (rank : 25) | θ value | 3.76295e-14 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
GIT2_HUMAN
|
||||||
| NC score | 0.548953 (rank : 26) | θ value | 2.79066e-09 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.544517 (rank : 27) | θ value | 8.11959e-09 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.536609 (rank : 28) | θ value | 6.21693e-09 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.534963 (rank : 29) | θ value | 1.38499e-08 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.531283 (rank : 30) | θ value | 2.36244e-08 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| NC score | 0.530237 (rank : 31) | θ value | 2.36244e-08 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.526127 (rank : 32) | θ value | 1.09739e-05 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.522131 (rank : 33) | θ value | 4.92598e-06 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.520514 (rank : 34) | θ value | 1.80886e-08 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.520161 (rank : 35) | θ value | 5.62301e-10 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.517091 (rank : 36) | θ value | 3.29651e-10 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.412114 (rank : 37) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.392406 (rank : 38) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.112508 (rank : 39) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
CT086_HUMAN
|
||||||
| NC score | 0.104613 (rank : 40) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.102542 (rank : 41) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.099813 (rank : 42) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.099324 (rank : 43) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_MOUSE
|
||||||
| NC score | 0.096975 (rank : 44) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.096860 (rank : 45) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
DP13B_HUMAN
|
||||||
| NC score | 0.088011 (rank : 46) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.086928 (rank : 47) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.085847 (rank : 48) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
ANR46_HUMAN
|
||||||
| NC score | 0.084635 (rank : 49) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.083862 (rank : 50) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
ANR46_MOUSE
|
||||||
| NC score | 0.083718 (rank : 51) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTZ5, Q1LZK9, Q8BV97 | Gene names | Ankrd46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR37_MOUSE
|
||||||
| NC score | 0.082316 (rank : 52) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569N2, Q8CHS0 | Gene names | Ankrd37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37. | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.077296 (rank : 53) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
ANR39_MOUSE
|
||||||
| NC score | 0.076881 (rank : 54) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D2X0 | Gene names | Ankrd39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR39_HUMAN
|
||||||
| NC score | 0.075641 (rank : 55) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q53RE8, Q59FU2, Q8N5X5, Q9P0S5 | Gene names | ANKRD39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
GABP3_MOUSE
|
||||||
| NC score | 0.073293 (rank : 56) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P81069 | Gene names | Gabpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GA-binding protein beta-2-1 chain (GABP subunit beta-2-1) (GABPB2-1). | |||||
|
ANS4B_MOUSE
|
||||||
| NC score | 0.072978 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.072256 (rank : 58) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| NC score | 0.071699 (rank : 59) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
ANR37_HUMAN
|
||||||
| NC score | 0.071320 (rank : 60) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z713 | Gene names | ANKRD37, LPR2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37 (Low-density lipoprotein receptor-related protein 2-binding protein) (hLrp2bp). | |||||
|
MTPN_MOUSE
|
||||||
| NC score | 0.068730 (rank : 61) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62774, P80144, Q543M6, Q9DCN8 | Gene names | Mtpn, Gcdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotrophin (Protein V-1) (Granule cell differentiation protein). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.067961 (rank : 62) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.066998 (rank : 63) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
MTPN_HUMAN
|
||||||
| NC score | 0.066869 (rank : 64) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58546 | Gene names | MTPN | |||
|
Domain Architecture |

|
|||||
| Description | Myotrophin (Protein V-1). | |||||
|
ANS4B_HUMAN
|
||||||
| NC score | 0.066524 (rank : 65) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.066227 (rank : 66) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.066185 (rank : 67) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CDN2D_HUMAN
|
||||||
| NC score | 0.065611 (rank : 68) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.065207 (rank : 69) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
ANKR2_HUMAN
|
||||||
| NC score | 0.063811 (rank : 70) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.063688 (rank : 71) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.062822 (rank : 72) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.061633 (rank : 73) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.061543 (rank : 74) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
USH1G_MOUSE
|
||||||
| NC score | 0.061455 (rank : 75) | θ value | θ > 10 (rank : 198) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.061298 (rank : 76) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
USH1G_HUMAN
|
||||||
| NC score | 0.061185 (rank : 77) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.060673 (rank : 78) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.060581 (rank : 79) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ANR33_HUMAN
|
||||||
| NC score | 0.060233 (rank : 80) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR10_MOUSE
|
||||||
| NC score | 0.059999 (rank : 81) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.059843 (rank : 82) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.059840 (rank : 83) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.059782 (rank : 84) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.059756 (rank : 85) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.059739 (rank : 86) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.059360 (rank : 87) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.059339 (rank : 88) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.058981 (rank : 89) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.058953 (rank : 90) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.058852 (rank : 91) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.058779 (rank : 92) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
ANKR2_MOUSE
|
||||||
| NC score | 0.058770 (rank : 93) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.058767 (rank : 94) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.058743 (rank : 95) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.058599 (rank : 96) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.058513 (rank : 97) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.058340 (rank : 98) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
ANR23_HUMAN
|
||||||
| NC score | 0.058109 (rank : 99) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.057900 (rank : 100) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| NC score | 0.057449 (rank : 101) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.057046 (rank : 102) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.056748 (rank : 103) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.056406 (rank : 104) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.056341 (rank : 105) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.056280 (rank : 106) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.056206 (rank : 107) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.055739 (rank : 108) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ACBD6_HUMAN
|
||||||
| NC score | 0.055620 (rank : 109) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.055576 (rank : 110) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.055218 (rank : 111) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.055198 (rank : 112) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.055114 (rank : 113) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.054595 (rank : 114) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.054571 (rank : 115) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
ACBD6_MOUSE
|
||||||
| NC score | 0.054450 (rank : 116) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.054285 (rank : 117) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.054183 (rank : 118) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.054127 (rank : 119) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
ANR49_HUMAN
|
||||||
| NC score | 0.053927 (rank : 120) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.053804 (rank : 121) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ANKR5_HUMAN
|
||||||
| NC score | 0.053797 (rank : 122) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NU02, Q9H6Y9 | Gene names | ANKRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.053644 (rank : 123) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PP16A_MOUSE
|
||||||
| NC score | 0.053557 (rank : 124) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.053485 (rank : 125) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
ANKR5_MOUSE
|
||||||
| NC score | 0.053415 (rank : 126) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.053400 (rank : 127) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ASB16_HUMAN
|
||||||
| NC score | 0.053094 (rank : 128) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB7_MOUSE
|
||||||
| NC score | 0.053001 (rank : 129) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_HUMAN
|
||||||
| NC score | 0.053000 (rank : 130) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.052805 (rank : 131) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.052804 (rank : 132) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.052429 (rank : 133) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ASB12_MOUSE
|
||||||
| NC score | 0.052307 (rank : 134) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D738 | Gene names | Asb12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 12 (ASB-12). | |||||
|
ASB2_HUMAN
|
||||||
| NC score | 0.052229 (rank : 135) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
CD3E_MOUSE
|
||||||
| NC score | 0.052163 (rank : 136) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22646 | Gene names | Cd3e | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD3 epsilon chain precursor (T-cell surface antigen T3/Leu-4 epsilon chain). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.052073 (rank : 137) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
AN13B_MOUSE
|
||||||
| NC score | 0.051969 (rank : 138) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5F259, Q6IR40 | Gene names | Ankrd13b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.051965 (rank : 139) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
AN36A_HUMAN
|
||||||
| NC score | 0.051944 (rank : 140) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
AN13B_HUMAN
|
||||||
| NC score | 0.051892 (rank : 141) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YJ7, Q8N7S9 | Gene names | ANKRD13B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
ASB16_MOUSE
|
||||||
| NC score | 0.051862 (rank : 142) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB12_HUMAN
|
||||||
| NC score | 0.051806 (rank : 143) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXK4, Q52LK4, Q6ISF9 | Gene names | ASB12 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 12 (ASB-12). | |||||
|
ANR15_HUMAN
|
||||||
| NC score | 0.051553 (rank : 144) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.051392 (rank : 145) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.051346 (rank : 146) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ANR49_MOUSE
|
||||||
| NC score | 0.051127 (rank : 147) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
PP16B_MOUSE
|
||||||
| NC score | 0.050823 (rank : 148) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHQ3 | Gene names | Ppp1r16b | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory inhibitor subunit 16B (TGF-beta- inhibited membrane-associated protein) (CAAX box protein TIMAP). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.050688 (rank : 149) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.050604 (rank : 150) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.050586 (rank : 151) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.050544 (rank : 152) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.050405 (rank : 153) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKS3_MOUSE
|
||||||
| NC score | 0.050385 (rank : 154) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.050306 (rank : 155) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
POTE8_HUMAN
|
||||||
| NC score | 0.050190 (rank : 156) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6S8J7, Q6S8J6 | Gene names | POTE8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 8. | |||||
|
ASB13_MOUSE
|
||||||
| NC score | 0.050111 (rank : 157) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VBX0 | Gene names | Asb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.050019 (rank : 158) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.045051 (rank : 159) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.039496 (rank : 160) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.032333 (rank : 161) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.032253 (rank : 162) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
ALKB5_HUMAN
|
||||||
| NC score | 0.031316 (rank : 163) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P6C2, Q9NXD6 | Gene names | ALKBH5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkylated repair protein alkB homolog 5. | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.025425 (rank : 164) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.024309 (rank : 165) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.021564 (rank : 166) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.019654 (rank : 167) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.018909 (rank : 168) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.018487 (rank : 169) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.018451 (rank : 170) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.017990 (rank : 171) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PRDM8_MOUSE
|
||||||
| NC score | 0.017570 (rank : 172) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ97 | Gene names | Prdm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.017390 (rank : 173) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.017245 (rank : 174) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.016984 (rank : 175) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.016890 (rank : 176) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.014916 (rank : 177) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.014673 (rank : 178) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
CCDC4_HUMAN
|
||||||
| NC score | 0.014673 (rank : 179) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.014188 (rank : 180) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.013751 (rank : 181) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.013396 (rank : 182) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.013376 (rank : 183) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.012286 (rank : 184) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
CLC6A_HUMAN
|
||||||
| NC score | 0.011307 (rank : 185) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
IRF7_HUMAN
|
||||||
| NC score | 0.010297 (rank : 186) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.009546 (rank : 187) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.008451 (rank : 188) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.007488 (rank : 189) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.005680 (rank : 190) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.005551 (rank : 191) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
INSM1_MOUSE
|
||||||
| NC score | 0.004723 (rank : 192) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
CSTN1_MOUSE
|
||||||
| NC score | 0.003564 (rank : 193) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
FOXC2_MOUSE
|
||||||
| NC score | 0.003552 (rank : 194) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61850, P97948, Q63869, Q8C694 | Gene names | Foxc2, Fkh14, Fkhl14, Mfh1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.002474 (rank : 195) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SALL2_MOUSE
|
||||||
| NC score | -0.000433 (rank : 196) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | -0.001156 (rank : 197) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ZN524_HUMAN
|
||||||
| NC score | -0.003131 (rank : 198) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96C55, Q6NW31, Q96IL7 | Gene names | ZNF524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 524. | |||||