Please be patient as the page loads
|
CSTN1_MOUSE
|
||||||
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |
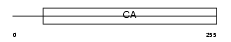
|
|||||
| Description | Calsyntenin-1 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CSTN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996671 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 159 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.981297 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |
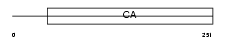
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.980361 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |
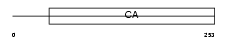
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.975721 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9BQT9, O94831 | Gene names | CLSTN3, CS3, KIAA0726 | |||
|
Domain Architecture |
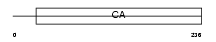
|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
CSTN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.972256 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q99JH7 | Gene names | Clstn3, Cs3, Cstn3 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
FATH_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 7) | NC score | 0.506596 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 8) | NC score | 0.561220 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 9) | NC score | 0.560796 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 10) | NC score | 0.486634 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
PCDB8_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 11) | NC score | 0.532640 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN66 | Gene names | PCDHB8, PCDH3I | |||
|
Domain Architecture |
|
|||||
| Description | Protocadherin beta 8 precursor (PCDH-beta8) (Protocadherin 3I). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 12) | NC score | 0.485819 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
PCD19_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 13) | NC score | 0.537296 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 14) | NC score | 0.480281 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 15) | NC score | 0.486358 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 16) | NC score | 0.477922 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
PCD19_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 17) | NC score | 0.535494 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCDBD_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 18) | NC score | 0.527937 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F0 | Gene names | PCDHB13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 13 precursor (PCDH-beta13). | |||||
|
PCDGJ_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.521964 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 20) | NC score | 0.474377 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
PCDGI_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 21) | NC score | 0.528300 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9Y5F9, Q9Y5C5 | Gene names | PCDHGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B6 precursor (PCDH-gamma-B6). | |||||
|
CAD26_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 22) | NC score | 0.559318 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 23) | NC score | 0.567299 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
CADH5_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 24) | NC score | 0.555871 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P33151 | Gene names | CDH5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (7B4 antigen) (CD144 antigen). | |||||
|
PCDGG_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 25) | NC score | 0.535922 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN71, O15099, Q9UN64 | Gene names | PCDHGB4, CDH20, FIB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B4 precursor (PCDH-gamma-B4) (Cadherin-20) (Fibroblast cadherin 2). | |||||
|
PCDH1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 26) | NC score | 0.551192 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 27) | NC score | 0.537454 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
PCDB7_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 28) | NC score | 0.522619 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E2 | Gene names | PCDHB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 7 precursor (PCDH-beta7). | |||||
|
CAD23_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 29) | NC score | 0.557908 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCDGH_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 30) | NC score | 0.527970 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
CAD12_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 31) | NC score | 0.544519 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
PCD15_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 32) | NC score | 0.566716 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 33) | NC score | 0.559319 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
CADH1_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 34) | NC score | 0.533085 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
CADH5_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 35) | NC score | 0.551250 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55284, O35542 | Gene names | Cdh5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (CD144 antigen). | |||||
|
DSG1A_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 36) | NC score | 0.520636 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q61495, Q8CE03 | Gene names | Dsg1a, Dsg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 alpha precursor (Dsg1-alpha) (Desmoglein-1) (Desmosomal glycoprotein I) (DG1) (DGI). | |||||
|
DSG1C_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 37) | NC score | 0.522594 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
PCD20_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 38) | NC score | 0.558644 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
PCDGL_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 39) | NC score | 0.530241 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F7, Q9Y5C3 | Gene names | PCDHGC4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C4 precursor (PCDH-gamma-C4). | |||||
|
CAD11_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 40) | NC score | 0.537320 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CADH6_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 41) | NC score | 0.534294 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
DSG1B_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 42) | NC score | 0.520040 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TSF1, Q7TQ60 | Gene names | Dsg1b, Dsg5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 beta precursor (Dsg1-beta) (Desmoglein-5). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 43) | NC score | 0.533536 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDBC_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 44) | NC score | 0.530745 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||
|
CAD23_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 45) | NC score | 0.557083 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCDB9_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 46) | NC score | 0.523839 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E1, Q9NRJ8 | Gene names | PCDHB9, PCDH3H | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 9 precursor (PCDH-beta9) (Protocadherin 3H). | |||||
|
CAD18_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 47) | NC score | 0.539752 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q13634 | Gene names | CDH18, CDH14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-18 precursor (Cadherin-14). | |||||
|
PCDA2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 48) | NC score | 0.514522 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDBB_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 49) | NC score | 0.528972 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F2 | Gene names | PCDHB11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 11 precursor (PCDH-beta11). | |||||
|
PCDGE_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 50) | NC score | 0.516803 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G2, Q9UN65 | Gene names | PCDHGB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B2 precursor (PCDH-gamma-B2). | |||||
|
CADH1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 51) | NC score | 0.523180 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P12830, Q13799, Q14216, Q15855, Q16194, Q4PJ14 | Gene names | CDH1, CDHE, UVO | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (CAM 120/80) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
PCDAD_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 52) | NC score | 0.513033 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDB1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 53) | NC score | 0.529984 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F3 | Gene names | PCDHB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 1 precursor (PCDH-beta1). | |||||
|
CADH9_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 54) | NC score | 0.533519 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9ULB4 | Gene names | CDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-9 precursor. | |||||
|
PCD20_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 55) | NC score | 0.555935 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
CAD20_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 56) | NC score | 0.539702 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HBT6 | Gene names | CDH20, CDH7L3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-20 precursor. | |||||
|
CADH6_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 57) | NC score | 0.531900 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
PCDB5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 58) | NC score | 0.520650 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
PCDH7_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 59) | NC score | 0.550940 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
CADH3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 60) | NC score | 0.531901 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
DSG1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 61) | NC score | 0.515880 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q02413 | Gene names | DSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-1 precursor (Desmosomal glycoprotein 1) (DG1) (DGI) (Pemphigus foliaceus antigen). | |||||
|
PCD12_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 62) | NC score | 0.530007 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDBF_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 63) | NC score | 0.524404 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E8, Q8IUX5 | Gene names | PCDHB15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 15 precursor (PCDH-beta15). | |||||
|
PCDGF_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 64) | NC score | 0.518522 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
PCDBG_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 65) | NC score | 0.523407 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NRJ7, Q8IYD5, Q96SE9, Q9HCF1 | Gene names | PCDHB16, KIAA1621, PCDH3X | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 16 precursor (PCDH-beta16) (Protocadherin 3X). | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 66) | NC score | 0.549154 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
DSC2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 67) | NC score | 0.497634 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
PCDGD_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 68) | NC score | 0.524073 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
CAD10_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 69) | NC score | 0.536586 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
DSG2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 70) | NC score | 0.513383 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
PCDB3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 71) | NC score | 0.522201 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
CAD11_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 72) | NC score | 0.535162 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CADH7_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 73) | NC score | 0.536134 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
DSG2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 74) | NC score | 0.525130 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
DSG4_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 75) | NC score | 0.519913 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q7TMD7 | Gene names | Dsg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
PCDB4_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 76) | NC score | 0.528128 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||
|
PCDBA_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 77) | NC score | 0.518415 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN67, Q96T99 | Gene names | PCDHB10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 10 precursor (PCDH-beta10). | |||||
|
PCDG1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 78) | NC score | 0.512859 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
PCDGK_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 79) | NC score | 0.520144 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
CAD19_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 80) | NC score | 0.545312 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H159 | Gene names | CDH19, CDH7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-19 precursor. | |||||
|
PCDB6_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 81) | NC score | 0.526531 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E3 | Gene names | PCDHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 6 precursor (PCDH-beta6). | |||||
|
PCDA1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 82) | NC score | 0.504325 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
CADH3_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 83) | NC score | 0.531604 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P10287, Q61465 | Gene names | Cdh3, Cdhp | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
PCDB2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 84) | NC score | 0.516972 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E7 | Gene names | PCDHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 2 precursor (PCDH-beta2). | |||||
|
PCDG7_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 85) | NC score | 0.517264 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDH8_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 86) | NC score | 0.519758 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O95206, Q8IYE9, Q96SF1 | Gene names | PCDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-8 precursor (Arcadlin). | |||||
|
PCDBE_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 87) | NC score | 0.519250 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E9 | Gene names | PCDHB14 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 14 precursor (PCDH-beta14). | |||||
|
PCDGA_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 88) | NC score | 0.509625 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
CAD22_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 89) | NC score | 0.527233 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
DSG3_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 90) | NC score | 0.519585 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
PCDLK_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 91) | NC score | 0.543404 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
CAD22_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 92) | NC score | 0.527365 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CAD26_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 93) | NC score | 0.542167 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
DSG4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 94) | NC score | 0.522570 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
PCDAB_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 95) | NC score | 0.505546 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
CAD24_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 96) | NC score | 0.533429 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
CADH8_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 97) | NC score | 0.532191 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55286, Q9ULB2 | Gene names | CDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CADH8_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 98) | NC score | 0.532119 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P97291 | Gene names | Cdh8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CAD15_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 99) | NC score | 0.512608 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P33146, Q9QYZ7 | Gene names | Cdh15, Cdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
PCDA6_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 100) | NC score | 0.500930 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
DSC3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 101) | NC score | 0.492809 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q14574, Q14200, Q9HAZ9 | Gene names | DSC3, DSC4 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor (Desmocollin-4) (HT-CP). | |||||
|
CAD15_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 102) | NC score | 0.501333 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
DSG3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 103) | NC score | 0.515050 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |
|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
PCDGB_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 104) | NC score | 0.504926 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
PCD10_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 105) | NC score | 0.521239 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||
|
PCDG8_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 106) | NC score | 0.511310 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G5, O15039 | Gene names | PCDHGA8, KIAA0327 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A8 precursor (PCDH-gamma-A8). | |||||
|
PCDG5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 107) | NC score | 0.505714 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
CAD16_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 108) | NC score | 0.530074 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | O88338, Q9JLZ5 | Gene names | Cdh16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
CADH4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 109) | NC score | 0.524011 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
PCD18_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 110) | NC score | 0.508668 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HCL0 | Gene names | PCDH18, KIAA1562 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-18 precursor. | |||||
|
PCDA4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 111) | NC score | 0.500861 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA8_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 112) | NC score | 0.503484 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDG9_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 113) | NC score | 0.509294 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G4, Q9Y5C9 | Gene names | PCDHGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A9 precursor (PCDH-gamma-A9). | |||||
|
CADH4_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 114) | NC score | 0.524549 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
PCDA3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 115) | NC score | 0.499897 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDG2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 116) | NC score | 0.506255 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
CADH2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 117) | NC score | 0.509623 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P19022, Q14923 | Gene names | CDH2, CDHN, NCAD | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
PCDAA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 118) | NC score | 0.491386 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDAC_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 119) | NC score | 0.499649 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDG4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 120) | NC score | 0.504329 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
CAD13_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 121) | NC score | 0.528255 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55290, Q8TBX3 | Gene names | CDH13, CDHH | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin) (P105). | |||||
|
CAD13_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 122) | NC score | 0.527930 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WTR5 | Gene names | Cdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin). | |||||
|
PCDC2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 123) | NC score | 0.508839 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
PCDG6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 124) | NC score | 0.505482 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||
|
PCDGC_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 125) | NC score | 0.505346 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
CADH2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 126) | NC score | 0.509312 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CAD17_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 127) | NC score | 0.521698 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 128) | NC score | 0.497304 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDG3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 129) | NC score | 0.496660 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
DSC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 130) | NC score | 0.484506 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
DSC3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 131) | NC score | 0.481639 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55850, O55110, O55122 | Gene names | Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor. | |||||
|
PCDC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 132) | NC score | 0.498515 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
DSC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 133) | NC score | 0.483366 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55849 | Gene names | Dsc1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor. | |||||
|
DSC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 134) | NC score | 0.482227 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P55292, Q64734 | Gene names | Dsc2, Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Epithelial type 2 desmocollin). | |||||
|
PCDA4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 135) | NC score | 0.493008 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 136) | NC score | 0.492026 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 137) | NC score | -0.000002 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CAD17_MOUSE
|
||||||
| θ value | 1.06291 (rank : 138) | NC score | 0.522276 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9R100 | Gene names | Cdh17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (BILL- cadherin) (P130). | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 139) | NC score | 0.051937 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 140) | NC score | 0.018330 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 141) | NC score | 0.016846 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 142) | NC score | 0.045960 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 143) | NC score | 0.493375 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
INAR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 144) | NC score | 0.017611 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48551 | Gene names | IFNAR2, IFNARB | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 2.36792 (rank : 145) | NC score | 0.018210 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 146) | NC score | 0.022161 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 147) | NC score | 0.004913 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 148) | NC score | 0.009408 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 149) | NC score | -0.004623 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN175_HUMAN
|
||||||
| θ value | 3.0926 (rank : 150) | NC score | -0.005344 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y473 | Gene names | ZNF175 | |||
|
Domain Architecture |
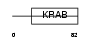
|
|||||
| Description | Zinc finger protein 175 (Zinc finger protein OTK18). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 151) | NC score | 0.035873 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 152) | NC score | 0.013321 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
DCR1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 153) | NC score | 0.014225 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 154) | NC score | 0.008125 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 155) | NC score | 0.017262 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 156) | NC score | -0.006791 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 157) | NC score | 0.008915 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 4.03905 (rank : 158) | NC score | 0.014599 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
GLTL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 159) | NC score | 0.003629 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 160) | NC score | 0.009889 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 161) | NC score | 0.025014 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 162) | NC score | 0.000785 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
Z297B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 163) | NC score | -0.003909 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 792 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43298 | Gene names | ZNF297B, KIAA0414, ZBTB22B | |||
|
Domain Architecture |
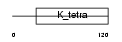
|
|||||
| Description | Zinc finger protein 297B (ZnF-x) (Zinc finger and BTB domain- containing protein 22B). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.007688 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | -0.003731 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | -0.005142 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
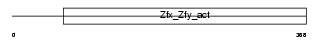
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.003564 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.003777 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.009701 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
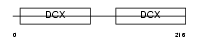
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
HXD10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | -0.002476 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28359, Q91WU9 | Gene names | Hoxd10, Hox-4.5, Hoxd-10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D10 (Hox-4.5) (Hox-5.3). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.004962 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.005923 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.004690 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.013519 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SIG11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.002322 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL6 | Gene names | SIGLEC11 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 11 precursor (Siglec-11) (Sialic acid-binding lectin 11). | |||||
|
CAD16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.487366 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
EMR4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.053200 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
GP133_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052889 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
GP144_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.053085 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.336703 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.363196 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PCD17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.508521 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||
|
PCDGM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.499573 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||
|
CSTN1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN1_HUMAN
|
||||||
| NC score | 0.996671 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 159 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN2_HUMAN
|
||||||
| NC score | 0.981297 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN2_MOUSE
|
||||||
| NC score | 0.980361 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN3_HUMAN
|
||||||
| NC score | 0.975721 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9BQT9, O94831 | Gene names | CLSTN3, CS3, KIAA0726 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
CSTN3_MOUSE
|
||||||
| NC score | 0.972256 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q99JH7 | Gene names | Clstn3, Cs3, Cstn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.567299 (rank : 7) | θ value | 5.81887e-07 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCD15_MOUSE
|
||||||
| NC score | 0.566716 (rank : 8) | θ value | 2.21117e-06 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.561220 (rank : 9) | θ value | 9.59137e-10 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.560796 (rank : 10) | θ value | 1.25267e-09 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.559319 (rank : 11) | θ value | 2.21117e-06 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
CAD26_HUMAN
|
||||||
| NC score | 0.559318 (rank : 12) | θ value | 5.81887e-07 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
PCD20_HUMAN
|
||||||
| NC score | 0.558644 (rank : 13) | θ value | 2.88788e-06 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
CAD23_MOUSE
|
||||||
| NC score | 0.557908 (rank : 14) | θ value | 1.69304e-06 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CAD23_HUMAN
|
||||||
| NC score | 0.557083 (rank : 15) | θ value | 4.92598e-06 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCD20_MOUSE
|
||||||
| NC score | 0.555935 (rank : 16) | θ value | 1.09739e-05 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
CADH5_HUMAN
|
||||||
| NC score | 0.555871 (rank : 17) | θ value | 9.92553e-07 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P33151 | Gene names | CDH5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (7B4 antigen) (CD144 antigen). | |||||
|
CADH5_MOUSE
|
||||||
| NC score | 0.551250 (rank : 18) | θ value | 2.88788e-06 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55284, O35542 | Gene names | Cdh5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (CD144 antigen). | |||||
|
PCDH1_HUMAN
|
||||||
| NC score | 0.551192 (rank : 19) | θ value | 9.92553e-07 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
PCDH7_HUMAN
|
||||||
| NC score | 0.550940 (rank : 20) | θ value | 1.43324e-05 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | 0.549154 (rank : 21) | θ value | 2.44474e-05 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
CAD19_HUMAN
|
||||||
| NC score | 0.545312 (rank : 22) | θ value | 7.1131e-05 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H159 | Gene names | CDH19, CDH7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-19 precursor. | |||||
|
CAD12_HUMAN
|
||||||
| NC score | 0.544519 (rank : 23) | θ value | 2.21117e-06 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
PCDLK_HUMAN
|
||||||
| NC score | 0.543404 (rank : 24) | θ value | 0.00020696 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
CAD26_MOUSE
|
||||||
| NC score | 0.542167 (rank : 25) | θ value | 0.000270298 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
CAD18_HUMAN
|
||||||
| NC score | 0.539752 (rank : 26) | θ value | 6.43352e-06 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q13634 | Gene names | CDH18, CDH14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-18 precursor (Cadherin-14). | |||||
|
CAD20_HUMAN
|
||||||
| NC score | 0.539702 (rank : 27) | θ value | 1.43324e-05 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HBT6 | Gene names | CDH20, CDH7L3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-20 precursor. | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.537454 (rank : 28) | θ value | 1.29631e-06 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
CAD11_HUMAN
|
||||||
| NC score | 0.537320 (rank : 29) | θ value | 3.77169e-06 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
PCD19_MOUSE
|
||||||
| NC score | 0.537296 (rank : 30) | θ value | 6.21693e-09 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
CAD10_HUMAN
|
||||||
| NC score | 0.536586 (rank : 31) | θ value | 4.1701e-05 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
CADH7_HUMAN
|
||||||
| NC score | 0.536134 (rank : 32) | θ value | 5.44631e-05 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
PCDGG_HUMAN
|
||||||
| NC score | 0.535922 (rank : 33) | θ value | 9.92553e-07 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN71, O15099, Q9UN64 | Gene names | PCDHGB4, CDH20, FIB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B4 precursor (PCDH-gamma-B4) (Cadherin-20) (Fibroblast cadherin 2). | |||||
|
PCD19_HUMAN
|
||||||
| NC score | 0.535494 (rank : 34) | θ value | 1.80886e-08 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
CAD11_MOUSE
|
||||||
| NC score | 0.535162 (rank : 35) | θ value | 5.44631e-05 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CADH6_HUMAN
|
||||||
| NC score | 0.534294 (rank : 36) | θ value | 3.77169e-06 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.533536 (rank : 37) | θ value | 3.77169e-06 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
CADH9_HUMAN
|
||||||
| NC score | 0.533519 (rank : 38) | θ value | 1.09739e-05 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9ULB4 | Gene names | CDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-9 precursor. | |||||
|
CAD24_HUMAN
|
||||||
| NC score | 0.533429 (rank : 39) | θ value | 0.00035302 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
CADH1_MOUSE
|
||||||
| NC score | 0.533085 (rank : 40) | θ value | 2.88788e-06 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
PCDB8_HUMAN
|
||||||
| NC score | 0.532640 (rank : 41) | θ value | 2.79066e-09 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN66 | Gene names | PCDHB8, PCDH3I | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 8 precursor (PCDH-beta8) (Protocadherin 3I). | |||||
|
CADH8_HUMAN
|
||||||
| NC score | 0.532191 (rank : 42) | θ value | 0.00035302 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55286, Q9ULB2 | Gene names | CDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CADH8_MOUSE
|
||||||
| NC score | 0.532119 (rank : 43) | θ value | 0.00035302 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P97291 | Gene names | Cdh8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CADH3_HUMAN
|
||||||
| NC score | 0.531901 (rank : 44) | θ value | 1.87187e-05 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CADH6_MOUSE
|
||||||
| NC score | 0.531900 (rank : 45) | θ value | 1.43324e-05 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CADH3_MOUSE
|
||||||
| NC score | 0.531604 (rank : 46) | θ value | 0.000121331 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P10287, Q61465 | Gene names | Cdh3, Cdhp | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
PCDBC_HUMAN
|
||||||
| NC score | 0.530745 (rank : 47) | θ value | 3.77169e-06 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||
|
PCDGL_HUMAN
|
||||||
| NC score | 0.530241 (rank : 48) | θ value | 2.88788e-06 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F7, Q9Y5C3 | Gene names | PCDHGC4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C4 precursor (PCDH-gamma-C4). | |||||
|
CAD16_MOUSE
|
||||||
| NC score | 0.530074 (rank : 49) | θ value | 0.00509761 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | O88338, Q9JLZ5 | Gene names | Cdh16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
PCD12_MOUSE
|
||||||
| NC score | 0.530007 (rank : 50) | θ value | 1.87187e-05 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDB1_HUMAN
|
||||||
| NC score | 0.529984 (rank : 51) | θ value | 8.40245e-06 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F3 | Gene names | PCDHB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 1 precursor (PCDH-beta1). | |||||
|
PCDBB_HUMAN
|
||||||
| NC score | 0.528972 (rank : 52) | θ value | 6.43352e-06 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F2 | Gene names | PCDHB11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 11 precursor (PCDH-beta11). | |||||
|
PCDGI_HUMAN
|
||||||
| NC score | 0.528300 (rank : 53) | θ value | 3.41135e-07 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9Y5F9, Q9Y5C5 | Gene names | PCDHGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B6 precursor (PCDH-gamma-B6). | |||||
|
CAD13_HUMAN
|
||||||
| NC score | 0.528255 (rank : 54) | θ value | 0.0148317 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55290, Q8TBX3 | Gene names | CDH13, CDHH | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin) (P105). | |||||
|
PCDB4_HUMAN
|
||||||
| NC score | 0.528128 (rank : 55) | θ value | 5.44631e-05 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||
|
PCDGH_HUMAN
|
||||||
| NC score | 0.527970 (rank : 56) | θ value | 1.69304e-06 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
PCDBD_HUMAN
|
||||||
| NC score | 0.527937 (rank : 57) | θ value | 1.53129e-07 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F0 | Gene names | PCDHB13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 13 precursor (PCDH-beta13). | |||||
|
CAD13_MOUSE
|
||||||
| NC score | 0.527930 (rank : 58) | θ value | 0.0148317 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WTR5 | Gene names | Cdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin). | |||||
|
CAD22_HUMAN
|
||||||
| NC score | 0.527365 (rank : 59) | θ value | 0.000270298 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CAD22_MOUSE
|
||||||
| NC score | 0.527233 (rank : 60) | θ value | 0.00020696 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
PCDB6_HUMAN
|
||||||
| NC score | 0.526531 (rank : 61) | θ value | 7.1131e-05 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E3 | Gene names | PCDHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 6 precursor (PCDH-beta6). | |||||
|
DSG2_HUMAN
|
||||||
| NC score | 0.525130 (rank : 62) | θ value | 5.44631e-05 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
CADH4_MOUSE
|
||||||
| NC score | 0.524549 (rank : 63) | θ value | 0.00665767 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
PCDBF_HUMAN
|
||||||
| NC score | 0.524404 (rank : 64) | θ value | 1.87187e-05 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E8, Q8IUX5 | Gene names | PCDHB15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 15 precursor (PCDH-beta15). | |||||
|
PCDGD_HUMAN
|
||||||
| NC score | 0.524073 (rank : 65) | θ value | 3.19293e-05 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
CADH4_HUMAN
|
||||||
| NC score | 0.524011 (rank : 66) | θ value | 0.00509761 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
PCDB9_HUMAN
|
||||||
| NC score | 0.523839 (rank : 67) | θ value | 4.92598e-06 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E1, Q9NRJ8 | Gene names | PCDHB9, PCDH3H | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 9 precursor (PCDH-beta9) (Protocadherin 3H). | |||||
|
PCDBG_HUMAN
|
||||||
| NC score | 0.523407 (rank : 68) | θ value | 2.44474e-05 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NRJ7, Q8IYD5, Q96SE9, Q9HCF1 | Gene names | PCDHB16, KIAA1621, PCDH3X | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 16 precursor (PCDH-beta16) (Protocadherin 3X). | |||||
|
CADH1_HUMAN
|
||||||
| NC score | 0.523180 (rank : 69) | θ value | 8.40245e-06 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P12830, Q13799, Q14216, Q15855, Q16194, Q4PJ14 | Gene names | CDH1, CDHE, UVO | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (CAM 120/80) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
PCDB7_HUMAN
|
||||||
| NC score | 0.522619 (rank : 70) | θ value | 1.29631e-06 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E2 | Gene names | PCDHB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 7 precursor (PCDH-beta7). | |||||
|
DSG1C_MOUSE
|
||||||
| NC score | 0.522594 (rank : 71) | θ value | 2.88788e-06 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
DSG4_HUMAN
|
||||||
| NC score | 0.522570 (rank : 72) | θ value | 0.000270298 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
CAD17_MOUSE
|
||||||
| NC score | 0.522276 (rank : 73) | θ value | 1.06291 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9R100 | Gene names | Cdh17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (BILL- cadherin) (P130). | |||||
|
PCDB3_HUMAN
|
||||||
| NC score | 0.522201 (rank : 74) | θ value | 4.1701e-05 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
PCDGJ_HUMAN
|
||||||
| NC score | 0.521964 (rank : 75) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
CAD17_HUMAN
|
||||||
| NC score | 0.521698 (rank : 76) | θ value | 0.0330416 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
PCD10_HUMAN
|
||||||
| NC score | 0.521239 (rank : 77) | θ value | 0.00228821 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||
|
PCDB5_HUMAN
|
||||||
| NC score | 0.520650 (rank : 78) | θ value | 1.43324e-05 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
DSG1A_MOUSE
|
||||||
| NC score | 0.520636 (rank : 79) | θ value | 2.88788e-06 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q61495, Q8CE03 | Gene names | Dsg1a, Dsg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 alpha precursor (Dsg1-alpha) (Desmoglein-1) (Desmosomal glycoprotein I) (DG1) (DGI). | |||||
|
PCDGK_HUMAN
|
||||||
| NC score | 0.520144 (rank : 80) | θ value | 5.44631e-05 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
DSG1B_MOUSE
|
||||||
| NC score | 0.520040 (rank : 81) | θ value | 3.77169e-06 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TSF1, Q7TQ60 | Gene names | Dsg1b, Dsg5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 beta precursor (Dsg1-beta) (Desmoglein-5). | |||||
|
DSG4_MOUSE
|
||||||
| NC score | 0.519913 (rank : 82) | θ value | 5.44631e-05 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q7TMD7 | Gene names | Dsg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
PCDH8_HUMAN
|
||||||
| NC score | 0.519758 (rank : 83) | θ value | 0.000121331 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O95206, Q8IYE9, Q96SF1 | Gene names | PCDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-8 precursor (Arcadlin). | |||||
|
DSG3_MOUSE
|
||||||
| NC score | 0.519585 (rank : 84) | θ value | 0.00020696 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
PCDBE_HUMAN
|
||||||
| NC score | 0.519250 (rank : 85) | θ value | 0.000158464 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E9 | Gene names | PCDHB14 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 14 precursor (PCDH-beta14). | |||||
|
PCDGF_HUMAN
|
||||||
| NC score | 0.518522 (rank : 86) | θ value | 1.87187e-05 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
PCDBA_HUMAN
|
||||||
| NC score | 0.518415 (rank : 87) | θ value | 5.44631e-05 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN67, Q96T99 | Gene names | PCDHB10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 10 precursor (PCDH-beta10). | |||||
|
PCDG7_HUMAN
|
||||||
| NC score | 0.517264 (rank : 88) | θ value | 0.000121331 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDB2_HUMAN
|
||||||
| NC score | 0.516972 (rank : 89) | θ value | 0.000121331 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5E7 | Gene names | PCDHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 2 precursor (PCDH-beta2). | |||||
|
PCDGE_HUMAN
|
||||||
| NC score | 0.516803 (rank : 90) | θ value | 6.43352e-06 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G2, Q9UN65 | Gene names | PCDHGB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B2 precursor (PCDH-gamma-B2). | |||||
|
DSG1_HUMAN
|
||||||
| NC score | 0.515880 (rank : 91) | θ value | 1.87187e-05 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q02413 | Gene names | DSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-1 precursor (Desmosomal glycoprotein 1) (DG1) (DGI) (Pemphigus foliaceus antigen). | |||||
|
DSG3_HUMAN
|
||||||
| NC score | 0.515050 (rank : 92) | θ value | 0.00102713 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
PCDA2_HUMAN
|
||||||
| NC score | 0.514522 (rank : 93) | θ value | 6.43352e-06 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
DSG2_MOUSE
|
||||||
| NC score | 0.513383 (rank : 94) | θ value | 4.1701e-05 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
PCDAD_HUMAN
|
||||||
| NC score | 0.513033 (rank : 95) | θ value | 8.40245e-06 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDG1_HUMAN
|
||||||
| NC score | 0.512859 (rank : 96) | θ value | 5.44631e-05 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
CAD15_MOUSE
|
||||||
| NC score | 0.512608 (rank : 97) | θ value | 0.000461057 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P33146, Q9QYZ7 | Gene names | Cdh15, Cdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
PCDG8_HUMAN
|
||||||
| NC score | 0.511310 (rank : 98) | θ value | 0.00298849 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G5, O15039 | Gene names | PCDHGA8, KIAA0327 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A8 precursor (PCDH-gamma-A8). | |||||
|
PCDGA_HUMAN
|
||||||
| NC score | 0.509625 (rank : 99) | θ value | 0.000158464 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
CADH2_HUMAN
|
||||||
| NC score | 0.509623 (rank : 100) | θ value | 0.0113563 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P19022, Q14923 | Gene names | CDH2, CDHN, NCAD | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CADH2_MOUSE
|
||||||
| NC score | 0.509312 (rank : 101) | θ value | 0.0252991 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
PCDG9_HUMAN
|
||||||
| NC score | 0.509294 (rank : 102) | θ value | 0.00509761 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G4, Q9Y5C9 | Gene names | PCDHGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A9 precursor (PCDH-gamma-A9). | |||||
|
PCDC2_HUMAN
|
||||||
| NC score | 0.508839 (rank : 103) | θ value | 0.0148317 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
PCD18_HUMAN
|
||||||
| NC score | 0.508668 (rank : 104) | θ value | 0.00509761 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HCL0 | Gene names | PCDH18, KIAA1562 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-18 precursor. | |||||
|
PCD17_HUMAN
|
||||||
| NC score | 0.508521 (rank : 105) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||
|
FATH_HUMAN
|
||||||
| NC score | 0.506596 (rank : 106) | θ value | 4.30538e-10 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
PCDG2_HUMAN
|
||||||
| NC score | 0.506255 (rank : 107) | θ value | 0.00869519 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
PCDG5_HUMAN
|
||||||
| NC score | 0.505714 (rank : 108) | θ value | 0.00390308 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
PCDAB_HUMAN
|
||||||
| NC score | 0.505546 (rank : 109) | θ value | 0.000270298 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDG6_HUMAN
|
||||||
| NC score | 0.505482 (rank : 110) | θ value | 0.0148317 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||
|
PCDGC_HUMAN
|
||||||
| NC score | 0.505346 (rank : 111) | θ value | 0.0148317 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
PCDGB_HUMAN
|
||||||
| NC score | 0.504926 (rank : 112) | θ value | 0.00134147 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
PCDG4_HUMAN
|
||||||
| NC score | 0.504329 (rank : 113) | θ value | 0.0113563 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
PCDA1_HUMAN
|
||||||
| NC score | 0.504325 (rank : 114) | θ value | 9.29e-05 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDA8_HUMAN
|
||||||
| NC score | 0.503484 (rank : 115) | θ value | 0.00509761 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
CAD15_HUMAN
|
||||||
| NC score | 0.501333 (rank : 116) | θ value | 0.00102713 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
PCDA6_HUMAN
|
||||||
| NC score | 0.500930 (rank : 117) | θ value | 0.000602161 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDA4_MOUSE
|
||||||
| NC score | 0.500861 (rank : 118) | θ value | 0.00509761 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA3_HUMAN
|
||||||
| NC score | 0.499897 (rank : 119) | θ value | 0.00665767 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDAC_HUMAN
|
||||||
| NC score | 0.499649 (rank : 120) | θ value | 0.0113563 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDGM_HUMAN
|
||||||
| NC score | 0.499573 (rank : 121) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||
|
PCDC1_HUMAN
|
||||||
| NC score | 0.498515 (rank : 122) | θ value | 0.163984 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
DSC2_HUMAN
|
||||||
| NC score | 0.497634 (rank : 123) | θ value | 3.19293e-05 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.497304 (rank : 124) | θ value | 0.0736092 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDG3_HUMAN
|
||||||
| NC score | 0.496660 (rank : 125) | θ value | 0.0736092 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.493375 (rank : 126) | θ value | 1.81305 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDA4_HUMAN
|
||||||
| NC score | 0.493008 (rank : 127) | θ value | 0.365318 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
DSC3_HUMAN
|
||||||
| NC score | 0.492809 (rank : 128) | θ value | 0.000786445 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q14574, Q14200, Q9HAZ9 | Gene names | DSC3, DSC4 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor (Desmocollin-4) (HT-CP). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.492026 (rank : 129) | θ value | 0.62314 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDAA_HUMAN
|
||||||
| NC score | 0.491386 (rank : 130) | θ value | 0.0113563 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
CAD16_HUMAN
|
||||||
| NC score | 0.487366 (rank : 131) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.486634 (rank : 132) | θ value | 1.63604e-09 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.486358 (rank : 133) | θ value | 8.11959e-09 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.485819 (rank : 134) | θ value | 3.64472e-09 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
DSC1_HUMAN
|
||||||
| NC score | 0.484506 (rank : 135) | θ value | 0.125558 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
DSC1_MOUSE
|
||||||
| NC score | 0.483366 (rank : 136) | θ value | 0.279714 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55849 | Gene names | Dsc1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor. | |||||
|
DSC2_MOUSE
|
||||||
| NC score | 0.482227 (rank : 137) | θ value | 0.279714 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P55292, Q64734 | Gene names | Dsc2, Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Epithelial type 2 desmocollin). | |||||
|
DSC3_MOUSE
|
||||||
| NC score | 0.481639 (rank : 138) | θ value | 0.163984 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55850, O55110, O55122 | Gene names | Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor. | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.480281 (rank : 139) | θ value | 8.11959e-09 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.477922 (rank : 140) | θ value | 1.80886e-08 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.474377 (rank : 141) | θ value | 2.61198e-07 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.363196 (rank : 142) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.336703 (rank : 143) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
EMR4_MOUSE
|
||||||
| NC score | 0.053200 (rank : 144) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.053085 (rank : 145) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.052889 (rank : 146) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.051937 (rank : 147) | θ value | 1.06291 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.045960 (rank : 148) | θ value | 1.81305 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.035873 (rank : 149) | θ value | 4.03905 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.025014 (rank : 150) | θ value | 5.27518 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.022161 (rank : 151) | θ value | 2.36792 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.018330 (rank : 152) | θ value | 1.06291 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.018210 (rank : 153) | θ value | 2.36792 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
INAR2_HUMAN
|
||||||
| NC score | 0.017611 (rank : 154) | θ value | 2.36792 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48551 | Gene names | IFNAR2, IFNARB | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.017262 (rank : 155) | θ value | 4.03905 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.016846 (rank : 156) | θ value | 1.06291 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.014599 (rank : 157) | θ value | 4.03905 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
DCR1A_HUMAN
|
||||||
| NC score | 0.014225 (rank : 158) | θ value | 4.03905 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.013519 (rank : 159) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.013321 (rank : 160) | θ value | 4.03905 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.009889 (rank : 161) | θ value | 5.27518 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.009701 (rank : 162) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.009408 (rank : 163) | θ value | 3.0926 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.008915 (rank : 164) | θ value | 4.03905 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.008125 (rank : 165) | θ value | 4.03905 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.007688 (rank : 166) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.005923 (rank : 167) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.004962 (rank : 168) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.004913 (rank : 169) | θ value | 3.0926 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.004690 (rank : 170) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.003777 (rank : 171) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
GLTL2_MOUSE
|
||||||
| NC score | 0.003629 (rank : 172) | θ value | 5.27518 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.003564 (rank : 173) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
SIG11_HUMAN
|
||||||
| NC score | 0.002322 (rank : 174) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL6 | Gene names | SIGLEC11 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 11 precursor (Siglec-11) (Sialic acid-binding lectin 11). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.000785 (rank : 175) | θ value | 5.27518 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | -0.000002 (rank : 176) | θ value | 0.813845 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
HXD10_MOUSE
|
||||||
| NC score | -0.002476 (rank : 177) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28359, Q91WU9 | Gene names | Hoxd10, Hox-4.5, Hoxd-10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D10 (Hox-4.5) (Hox-5.3). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | -0.003731 (rank : 178) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
Z297B_HUMAN
|
||||||
| NC score | -0.003909 (rank : 179) | θ value | 5.27518 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 792 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43298 | Gene names | ZNF297B, KIAA0414, ZBTB22B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297B (ZnF-x) (Zinc finger and BTB domain- containing protein 22B). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | -0.004623 (rank : 180) | θ value | 3.0926 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | -0.005142 (rank : 181) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
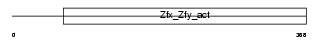
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZN175_HUMAN
|
||||||
| NC score | -0.005344 (rank : 182) | θ value | 3.0926 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y473 | Gene names | ZNF175 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 175 (Zinc finger protein OTK18). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | -0.006791 (rank : 183) | θ value | 4.03905 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||