Please be patient as the page loads
|
PCBP2_MOUSE
|
||||||
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PCBP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999586 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 5.94893e-161 (rank : 3) | NC score | 0.996271 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 5.94893e-161 (rank : 4) | NC score | 0.996271 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 1.20704e-137 (rank : 5) | NC score | 0.988013 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 1.20704e-137 (rank : 6) | NC score | 0.988027 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 2.72037e-97 (rank : 7) | NC score | 0.979568 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 1.76329e-96 (rank : 8) | NC score | 0.977849 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 1.42001e-37 (rank : 9) | NC score | 0.864804 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 1.42001e-37 (rank : 10) | NC score | 0.864804 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 11) | NC score | 0.779275 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 12) | NC score | 0.658079 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 13) | NC score | 0.773170 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 14) | NC score | 0.601141 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 15) | NC score | 0.604083 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 16) | NC score | 0.611453 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 17) | NC score | 0.627118 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 18) | NC score | 0.610377 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.456225 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 20) | NC score | 0.453831 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.378367 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 22) | NC score | 0.378487 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.021228 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.215765 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.201855 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.104066 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
SAM68_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.138046 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.136039 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.044386 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.057891 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.018234 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
EGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | -0.000769 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
CSTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.009408 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |
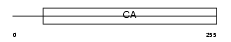
|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.005972 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.012480 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.014731 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.010824 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.010186 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.098598 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.096391 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.005119 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
SON_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.009087 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
UBXD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.024439 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |
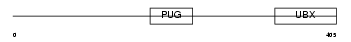
|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.054409 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.011170 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
SULF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.004435 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K007, Q8BLJ0, Q8C1D3 | Gene names | Sulf1 | |||
|
Domain Architecture |
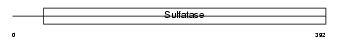
|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (MSulf-1). | |||||
|
WT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | -0.002265 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||
|
FMR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.063587 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.062378 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.212121 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.999586 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.996271 (rank : 3) | θ value | 5.94893e-161 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.996271 (rank : 4) | θ value | 5.94893e-161 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.988027 (rank : 5) | θ value | 1.20704e-137 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.988013 (rank : 6) | θ value | 1.20704e-137 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.979568 (rank : 7) | θ value | 2.72037e-97 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.977849 (rank : 8) | θ value | 1.76329e-96 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.864804 (rank : 9) | θ value | 1.42001e-37 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.864804 (rank : 10) | θ value | 1.42001e-37 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.779275 (rank : 11) | θ value | 3.17815e-21 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.773170 (rank : 12) | θ value | 2.88119e-14 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.658079 (rank : 13) | θ value | 3.40345e-15 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.627118 (rank : 14) | θ value | 5.43371e-13 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.611453 (rank : 15) | θ value | 5.43371e-13 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.610377 (rank : 16) | θ value | 7.09661e-13 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.604083 (rank : 17) | θ value | 4.16044e-13 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.601141 (rank : 18) | θ value | 1.42992e-13 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.456225 (rank : 19) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.453831 (rank : 20) | θ value | 4.45536e-07 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.378487 (rank : 21) | θ value | 8.40245e-06 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.378367 (rank : 22) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.215765 (rank : 23) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.212121 (rank : 24) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.201855 (rank : 25) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
SAM68_HUMAN
|
||||||
| NC score | 0.138046 (rank : 26) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| NC score | 0.136039 (rank : 27) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.104066 (rank : 28) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.098598 (rank : 29) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.096391 (rank : 30) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.063587 (rank : 31) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.062378 (rank : 32) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.057891 (rank : 33) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.054409 (rank : 34) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.044386 (rank : 35) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
UBXD1_HUMAN
|
||||||
| NC score | 0.024439 (rank : 36) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.021228 (rank : 37) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.018234 (rank : 38) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.014731 (rank : 39) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.012480 (rank : 40) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.011170 (rank : 41) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.010824 (rank : 42) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.010186 (rank : 43) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CSTN1_MOUSE
|
||||||
| NC score | 0.009408 (rank : 44) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.009087 (rank : 45) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.005972 (rank : 46) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.005119 (rank : 47) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
SULF1_MOUSE
|
||||||
| NC score | 0.004435 (rank : 48) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K007, Q8BLJ0, Q8C1D3 | Gene names | Sulf1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (MSulf-1). | |||||
|
EGFR_HUMAN
|
||||||
| NC score | -0.000769 (rank : 49) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
WT1_HUMAN
|
||||||
| NC score | -0.002265 (rank : 50) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||