Please be patient as the page loads
|
NOVA2_HUMAN
|
||||||
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NOVA2_HUMAN
|
||||||
| θ value | 7.8238e-137 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 6.90186e-109 (rank : 2) | NC score | 0.983298 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 3) | NC score | 0.767276 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 4) | NC score | 0.767398 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 5) | NC score | 0.766452 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 6) | NC score | 0.773170 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 7) | NC score | 0.769322 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 8) | NC score | 0.654394 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 9) | NC score | 0.769054 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 10) | NC score | 0.769054 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 11) | NC score | 0.772494 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 12) | NC score | 0.683494 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 13) | NC score | 0.680726 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 14) | NC score | 0.592683 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 15) | NC score | 0.592216 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 16) | NC score | 0.722887 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 17) | NC score | 0.722887 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 18) | NC score | 0.707716 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 19) | NC score | 0.633682 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 20) | NC score | 0.661988 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 21) | NC score | 0.269388 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 22) | NC score | 0.368032 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 23) | NC score | 0.365095 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.272156 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.284761 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
PNPT1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.104832 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
SAM68_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.168973 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.166275 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
PNPT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.111803 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.049846 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CN093_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.047008 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2W9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf93 homolog precursor. | |||||
|
FMR1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.133491 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.131055 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.155986 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.053910 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.156871 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.045774 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.056419 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.045818 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.146331 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | -0.001481 (rank : 56) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
NKX25_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.004253 (rank : 53) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42582, P97335 | Gene names | Nkx2-5, Csx, Nkx-2.5, Nkx2e | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.5 (Homeobox protein NK-2 homolog E) (Cardiac- specific homeobox) (Homeobox protein CSX). | |||||
|
CDCA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.018700 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FF9 | Gene names | CDCA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sororin (Cell division cycle-associated protein 5) (p35). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.010357 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | -0.001473 (rank : 55) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
LRFN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.001621 (rank : 54) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PJG9, Q9BWJ0 | Gene names | LRFN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 4 precursor. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.007029 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SHD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.010827 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.007160 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.005400 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
MYO3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | -0.003181 (rank : 57) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
FXR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.092285 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.069061 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.058660 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
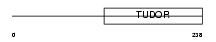
TDRD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.086827 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.062907 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.064658 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.8238e-137 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.983298 (rank : 2) | θ value | 6.90186e-109 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.773170 (rank : 3) | θ value | 2.88119e-14 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.772494 (rank : 4) | θ value | 2.43908e-13 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.769322 (rank : 5) | θ value | 2.88119e-14 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.769054 (rank : 6) | θ value | 1.86753e-13 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.769054 (rank : 7) | θ value | 1.86753e-13 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.767398 (rank : 8) | θ value | 1.92812e-18 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.767276 (rank : 9) | θ value | 1.92812e-18 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.766452 (rank : 10) | θ value | 4.44505e-15 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.722887 (rank : 11) | θ value | 4.59992e-12 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.722887 (rank : 12) | θ value | 4.59992e-12 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.707716 (rank : 13) | θ value | 1.02475e-11 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.683494 (rank : 14) | θ value | 4.16044e-13 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.680726 (rank : 15) | θ value | 7.09661e-13 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.661988 (rank : 16) | θ value | 1.38499e-08 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.654394 (rank : 17) | θ value | 3.76295e-14 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.633682 (rank : 18) | θ value | 1.74796e-11 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.592683 (rank : 19) | θ value | 1.58096e-12 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.592216 (rank : 20) | θ value | 2.69671e-12 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.368032 (rank : 21) | θ value | 0.0113563 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.365095 (rank : 22) | θ value | 0.0193708 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.284761 (rank : 23) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.272156 (rank : 24) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.269388 (rank : 25) | θ value | 0.00509761 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
SAM68_HUMAN
|
||||||
| NC score | 0.168973 (rank : 26) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| NC score | 0.166275 (rank : 27) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.156871 (rank : 28) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.155986 (rank : 29) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.146331 (rank : 30) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.133491 (rank : 31) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.131055 (rank : 32) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
PNPT1_HUMAN
|
||||||
| NC score | 0.111803 (rank : 33) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
PNPT1_MOUSE
|
||||||
| NC score | 0.104832 (rank : 34) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.092285 (rank : 35) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.086827 (rank : 36) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.069061 (rank : 37) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD7_MOUSE
|
||||||
| NC score | 0.064658 (rank : 38) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.062907 (rank : 39) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.058660 (rank : 40) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.056419 (rank : 41) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.053910 (rank : 42) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.049846 (rank : 43) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CN093_MOUSE
|
||||||
| NC score | 0.047008 (rank : 44) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2W9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf93 homolog precursor. | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.045818 (rank : 45) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.045774 (rank : 46) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
CDCA5_HUMAN
|
||||||
| NC score | 0.018700 (rank : 47) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FF9 | Gene names | CDCA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sororin (Cell division cycle-associated protein 5) (p35). | |||||
|
SHD_HUMAN
|
||||||
| NC score | 0.010827 (rank : 48) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.010357 (rank : 49) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.007160 (rank : 50) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.007029 (rank : 51) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.005400 (rank : 52) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
NKX25_MOUSE
|
||||||
| NC score | 0.004253 (rank : 53) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42582, P97335 | Gene names | Nkx2-5, Csx, Nkx-2.5, Nkx2e | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.5 (Homeobox protein NK-2 homolog E) (Cardiac- specific homeobox) (Homeobox protein CSX). | |||||
|
LRFN4_HUMAN
|
||||||
| NC score | 0.001621 (rank : 54) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PJG9, Q9BWJ0 | Gene names | LRFN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 4 precursor. | |||||
|
EGR2_HUMAN
|
||||||
| NC score | -0.001473 (rank : 55) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.001481 (rank : 56) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
MYO3A_HUMAN
|
||||||
| NC score | -0.003181 (rank : 57) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||