Please be patient as the page loads
|
FUBP3_HUMAN
|
||||||
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FUBP3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 3.16172e-154 (rank : 2) | NC score | 0.961939 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 2.04937e-153 (rank : 3) | NC score | 0.961568 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 7.57797e-132 (rank : 4) | NC score | 0.957220 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 5) | NC score | 0.629728 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 6) | NC score | 0.655875 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 7) | NC score | 0.664398 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 8) | NC score | 0.655379 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 9) | NC score | 0.669695 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 10) | NC score | 0.669695 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 11) | NC score | 0.658079 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 12) | NC score | 0.612948 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 13) | NC score | 0.575036 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 14) | NC score | 0.572769 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 15) | NC score | 0.660505 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 16) | NC score | 0.654177 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 17) | NC score | 0.707716 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 18) | NC score | 0.670600 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 19) | NC score | 0.458971 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 20) | NC score | 0.458562 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 21) | NC score | 0.587083 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 22) | NC score | 0.587083 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 23) | NC score | 0.368053 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 24) | NC score | 0.200426 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
SAM68_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 25) | NC score | 0.195068 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 26) | NC score | 0.185300 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 27) | NC score | 0.198884 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 28) | NC score | 0.243925 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.232090 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 30) | NC score | 0.177857 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.030049 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.162048 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.061396 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.020906 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
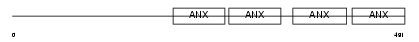
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
FMR1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.161986 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.022963 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TM55B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.028793 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.066691 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.007527 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.011287 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.070654 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.074264 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.017627 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.020354 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.020957 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.020676 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.036545 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.028874 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.020334 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
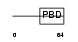
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.012629 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.015726 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.010173 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.016314 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PNPT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.042170 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.018374 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
HME1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.004296 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |
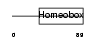
|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.012331 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.034551 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ERG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.010209 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.005403 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PLK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | -0.003211 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
PNPT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.047056 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.008717 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | -0.000309 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.009329 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.001706 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.002833 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.002831 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
FXR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.114424 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
KHDR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056175 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75525, Q6NUL8, Q9UPA8 | Gene names | KHDRBS3, SALP, SLM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 3 (Sam68-like mammalian protein 2) (SLM-2) (Sam68-like phosphotyrosine protein) (RNA-binding protein T-Star). | |||||
|
KHDR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.055847 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R226, O88624 | Gene names | Khdrbs3, Salp, Slm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 3 (Sam68-like mammalian protein 2) (SLM-2) (RNA-binding protein Etoile). | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.054776 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.068834 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
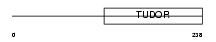
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051326 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.961939 (rank : 2) | θ value | 3.16172e-154 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.961568 (rank : 3) | θ value | 2.04937e-153 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.957220 (rank : 4) | θ value | 7.57797e-132 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.707716 (rank : 5) | θ value | 1.02475e-11 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.670600 (rank : 6) | θ value | 6.21693e-09 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.669695 (rank : 7) | θ value | 1.99529e-15 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.669695 (rank : 8) | θ value | 1.99529e-15 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.664398 (rank : 9) | θ value | 1.058e-16 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.660505 (rank : 10) | θ value | 1.58096e-12 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.658079 (rank : 11) | θ value | 3.40345e-15 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.655875 (rank : 12) | θ value | 6.20254e-17 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.655379 (rank : 13) | θ value | 1.058e-16 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.654177 (rank : 14) | θ value | 7.84624e-12 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.629728 (rank : 15) | θ value | 6.20254e-17 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.612948 (rank : 16) | θ value | 2.20605e-14 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.587083 (rank : 17) | θ value | 9.92553e-07 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.587083 (rank : 18) | θ value | 9.92553e-07 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.575036 (rank : 19) | θ value | 2.43908e-13 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.572769 (rank : 20) | θ value | 5.43371e-13 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.458971 (rank : 21) | θ value | 5.26297e-08 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.458562 (rank : 22) | θ value | 5.26297e-08 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.368053 (rank : 23) | θ value | 7.1131e-05 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.243925 (rank : 24) | θ value | 0.0113563 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.232090 (rank : 25) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.200426 (rank : 26) | θ value | 0.00390308 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.198884 (rank : 27) | θ value | 0.00665767 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
SAM68_HUMAN
|
||||||
| NC score | 0.195068 (rank : 28) | θ value | 0.00390308 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| NC score | 0.185300 (rank : 29) | θ value | 0.00509761 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.177857 (rank : 30) | θ value | 0.0193708 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.162048 (rank : 31) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.161986 (rank : 32) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.114424 (rank : 33) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.074264 (rank : 34) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.070654 (rank : 35) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.068834 (rank : 36) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.066691 (rank : 37) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.061396 (rank : 38) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
KHDR3_HUMAN
|
||||||
| NC score | 0.056175 (rank : 39) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75525, Q6NUL8, Q9UPA8 | Gene names | KHDRBS3, SALP, SLM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 3 (Sam68-like mammalian protein 2) (SLM-2) (Sam68-like phosphotyrosine protein) (RNA-binding protein T-Star). | |||||
|
KHDR3_MOUSE
|
||||||
| NC score | 0.055847 (rank : 40) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R226, O88624 | Gene names | Khdrbs3, Salp, Slm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 3 (Sam68-like mammalian protein 2) (SLM-2) (RNA-binding protein Etoile). | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.054776 (rank : 41) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD7_MOUSE
|
||||||
| NC score | 0.051326 (rank : 42) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
PNPT1_HUMAN
|
||||||
| NC score | 0.047056 (rank : 43) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
PNPT1_MOUSE
|
||||||
| NC score | 0.042170 (rank : 44) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.036545 (rank : 45) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.034551 (rank : 46) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.030049 (rank : 47) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.028874 (rank : 48) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
TM55B_HUMAN
|
||||||
| NC score | 0.028793 (rank : 49) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.022963 (rank : 50) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.020957 (rank : 51) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.020906 (rank : 52) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.020676 (rank : 53) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.020354 (rank : 54) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.020334 (rank : 55) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.018374 (rank : 56) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.017627 (rank : 57) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.016314 (rank : 58) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.015726 (rank : 59) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.012629 (rank : 60) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.012331 (rank : 61) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.011287 (rank : 62) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.010209 (rank : 63) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.010173 (rank : 64) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.009329 (rank : 65) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.008717 (rank : 66) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.007527 (rank : 67) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.005403 (rank : 68) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
HME1_MOUSE
|
||||||
| NC score | 0.004296 (rank : 69) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.002833 (rank : 70) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.002831 (rank : 71) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.001706 (rank : 72) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | -0.000309 (rank : 73) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PLK4_HUMAN
|
||||||
| NC score | -0.003211 (rank : 74) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||