Please be patient as the page loads
|
TDRKH_MOUSE
|
||||||
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TDRKH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993872 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 3) | NC score | 0.543585 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD5_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 4) | NC score | 0.551629 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD7_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 5) | NC score | 0.507060 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 6) | NC score | 0.498418 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 7) | NC score | 0.575036 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 8) | NC score | 0.555308 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 9) | NC score | 0.556019 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 10) | NC score | 0.592683 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 11) | NC score | 0.509250 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 12) | NC score | 0.539415 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 13) | NC score | 0.491036 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 14) | NC score | 0.463575 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 15) | NC score | 0.413135 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 16) | NC score | 0.557717 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 17) | NC score | 0.468006 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 18) | NC score | 0.468860 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 19) | NC score | 0.463270 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 20) | NC score | 0.463270 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 21) | NC score | 0.455145 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 22) | NC score | 0.453831 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 23) | NC score | 0.443521 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.445566 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
TDRD4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 25) | NC score | 0.342438 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.436084 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 27) | NC score | 0.436238 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
SND1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 28) | NC score | 0.282869 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 29) | NC score | 0.324720 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 30) | NC score | 0.248137 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 31) | NC score | 0.247395 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 32) | NC score | 0.319567 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 33) | NC score | 0.236366 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
SND1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 34) | NC score | 0.285122 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 35) | NC score | 0.272248 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 36) | NC score | 0.200466 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 37) | NC score | 0.409047 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 38) | NC score | 0.409047 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
FMR1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 39) | NC score | 0.219734 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 40) | NC score | 0.218248 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
STK31_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.077415 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BXU1, Q9BXH8 | Gene names | STK31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1) (Serine/threonine- protein kinase NYD-SPK). | |||||
|
NCF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.029439 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
HSF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.020679 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
IKBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.024460 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.008359 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.008360 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
HDGF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.019613 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.016539 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
THAP8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.028236 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.014673 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
FYN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.000628 (rank : 60) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
FYN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.000771 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.004500 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
STK31_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.068569 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MW1 | Gene names | Stk31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1). | |||||
|
YES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.000400 (rank : 61) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07947 | Gene names | YES1, YES | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||
|
FR1OP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.012804 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JX5, Q32P17, Q8BH91 | Gene names | Fgfr1op | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FGFR1 oncogene partner. | |||||
|
HCK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.003268 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1035 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08631, Q5T7K1, Q5T7K2, Q96CC0, Q9H5Y5, Q9NUA4, Q9UMJ5 | Gene names | HCK | |||
|
Domain Architecture |
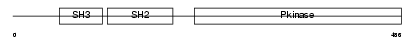
|
|||||
| Description | Tyrosine-protein kinase HCK (EC 2.7.10.2) (p59-HCK/p60-HCK) (Hemopoietic cell kinase). | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.016539 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.014829 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
SAM68_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.070407 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
SAM68_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.067794 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.993872 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.592683 (rank : 3) | θ value | 1.58096e-12 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.575036 (rank : 4) | θ value | 2.43908e-13 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.557717 (rank : 5) | θ value | 6.21693e-09 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.556019 (rank : 6) | θ value | 7.09661e-13 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.555308 (rank : 7) | θ value | 7.09661e-13 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.551629 (rank : 8) | θ value | 1.80466e-16 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.543585 (rank : 9) | θ value | 1.24977e-17 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.539415 (rank : 10) | θ value | 2.52405e-10 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.509250 (rank : 11) | θ value | 1.58096e-12 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD7_MOUSE
|
||||||
| NC score | 0.507060 (rank : 12) | θ value | 2.35696e-16 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.498418 (rank : 13) | θ value | 7.58209e-15 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.491036 (rank : 14) | θ value | 7.34386e-10 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.468860 (rank : 15) | θ value | 8.11959e-09 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.468006 (rank : 16) | θ value | 8.11959e-09 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.463575 (rank : 17) | θ value | 2.79066e-09 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.463270 (rank : 18) | θ value | 1.53129e-07 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.463270 (rank : 19) | θ value | 1.53129e-07 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.455145 (rank : 20) | θ value | 4.45536e-07 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.453831 (rank : 21) | θ value | 4.45536e-07 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.445566 (rank : 22) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.443521 (rank : 23) | θ value | 1.87187e-05 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.436238 (rank : 24) | θ value | 0.000270298 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.436084 (rank : 25) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.413135 (rank : 26) | θ value | 4.76016e-09 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.409047 (rank : 27) | θ value | 0.00869519 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.409047 (rank : 28) | θ value | 0.00869519 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
TDRD4_HUMAN
|
||||||
| NC score | 0.342438 (rank : 29) | θ value | 0.000158464 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.324720 (rank : 30) | θ value | 0.000270298 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.319567 (rank : 31) | θ value | 0.000786445 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
SND1_HUMAN
|
||||||
| NC score | 0.285122 (rank : 32) | θ value | 0.00102713 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
SND1_MOUSE
|
||||||
| NC score | 0.282869 (rank : 33) | θ value | 0.000270298 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.272248 (rank : 34) | θ value | 0.00228821 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.248137 (rank : 35) | θ value | 0.000786445 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.247395 (rank : 36) | θ value | 0.000786445 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.236366 (rank : 37) | θ value | 0.00102713 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.219734 (rank : 38) | θ value | 0.0148317 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.218248 (rank : 39) | θ value | 0.0148317 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.200466 (rank : 40) | θ value | 0.00390308 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
STK31_HUMAN
|
||||||
| NC score | 0.077415 (rank : 41) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BXU1, Q9BXH8 | Gene names | STK31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1) (Serine/threonine- protein kinase NYD-SPK). | |||||
|
SAM68_HUMAN
|
||||||
| NC score | 0.070407 (rank : 42) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
STK31_MOUSE
|
||||||
| NC score | 0.068569 (rank : 43) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MW1 | Gene names | Stk31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1). | |||||
|
SAM68_MOUSE
|
||||||
| NC score | 0.067794 (rank : 44) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
NCF2_HUMAN
|
||||||
| NC score | 0.029439 (rank : 45) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
THAP8_HUMAN
|
||||||
| NC score | 0.028236 (rank : 46) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.024460 (rank : 47) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.020679 (rank : 48) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
HDGF_HUMAN
|
||||||
| NC score | 0.019613 (rank : 49) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.016539 (rank : 50) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.016539 (rank : 51) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.014829 (rank : 52) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.014673 (rank : 53) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
FR1OP_MOUSE
|
||||||
| NC score | 0.012804 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JX5, Q32P17, Q8BH91 | Gene names | Fgfr1op | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FGFR1 oncogene partner. | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.008360 (rank : 55) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.008359 (rank : 56) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.004500 (rank : 57) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
HCK_HUMAN
|
||||||
| NC score | 0.003268 (rank : 58) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1035 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08631, Q5T7K1, Q5T7K2, Q96CC0, Q9H5Y5, Q9NUA4, Q9UMJ5 | Gene names | HCK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase HCK (EC 2.7.10.2) (p59-HCK/p60-HCK) (Hemopoietic cell kinase). | |||||
|
FYN_MOUSE
|
||||||
| NC score | 0.000771 (rank : 59) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
FYN_HUMAN
|
||||||
| NC score | 0.000628 (rank : 60) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
YES_HUMAN
|
||||||
| NC score | 0.000400 (rank : 61) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07947 | Gene names | YES1, YES | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||