Please be patient as the page loads
|
FMR1_HUMAN
|
||||||
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FMR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996288 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 2.511e-143 (rank : 3) | NC score | 0.972706 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 4.28312e-143 (rank : 4) | NC score | 0.972792 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 4.58675e-137 (rank : 5) | NC score | 0.962941 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 1.09426e-130 (rank : 6) | NC score | 0.962026 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.223077 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.219734 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.065166 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.152979 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.161986 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.105984 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.108081 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.151695 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.151879 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.100776 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.098331 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.133491 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.133581 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.013092 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SSF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.038933 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YU8 | Gene names | Ppan | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.010934 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.083747 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.083795 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.008404 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.016843 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.018467 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.010589 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.016902 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.001356 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SYS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.028871 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49591, Q9NSE3 | Gene names | SARS, SERS | |||
|
Domain Architecture |
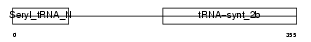
|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.069857 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.067614 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.029493 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ATS18_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.007497 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE60, Q6P4R5, Q6ZWJ9 | Gene names | ADAMTS18, ADAMTS21 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-18 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 18) (ADAM-TS 18) (ADAM-TS18). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.015580 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.020188 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ANKZ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.016061 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8Y5, Q9NVZ4 | Gene names | ANKZF1, ZNF744 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1 (Zinc finger protein 744). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.000824 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.118952 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.118952 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
SMD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.018013 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62314, P13641 | Gene names | SNRPD1 | |||
|
Domain Architecture |
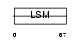
|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SMD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.018013 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62315, P13641, Q3UYM0 | Gene names | Snrpd1 | |||
|
Domain Architecture |
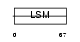
|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.002628 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SYS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.017637 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26638 | Gene names | Sars, Sars1, Sers | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.061266 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.061266 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.063723 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.063587 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.053995 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.054328 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.996288 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.972792 (rank : 3) | θ value | 4.28312e-143 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.972706 (rank : 4) | θ value | 2.511e-143 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.962941 (rank : 5) | θ value | 4.58675e-137 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.962026 (rank : 6) | θ value | 1.09426e-130 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.223077 (rank : 7) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.219734 (rank : 8) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.161986 (rank : 9) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.152979 (rank : 10) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.151879 (rank : 11) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.151695 (rank : 12) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.133581 (rank : 13) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.133491 (rank : 14) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.118952 (rank : 15) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.118952 (rank : 16) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.108081 (rank : 17) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.105984 (rank : 18) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.100776 (rank : 19) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.098331 (rank : 20) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.083795 (rank : 21) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.083747 (rank : 22) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.069857 (rank : 23) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.067614 (rank : 24) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.065166 (rank : 25) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.063723 (rank : 26) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.063587 (rank : 27) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.061266 (rank : 28) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.061266 (rank : 29) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.054328 (rank : 30) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.053995 (rank : 31) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
SSF1_MOUSE
|
||||||
| NC score | 0.038933 (rank : 32) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YU8 | Gene names | Ppan | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.029493 (rank : 33) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SYS_HUMAN
|
||||||
| NC score | 0.028871 (rank : 34) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49591, Q9NSE3 | Gene names | SARS, SERS | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.020188 (rank : 35) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.018467 (rank : 36) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SMD1_HUMAN
|
||||||
| NC score | 0.018013 (rank : 37) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62314, P13641 | Gene names | SNRPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SMD1_MOUSE
|
||||||
| NC score | 0.018013 (rank : 38) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62315, P13641, Q3UYM0 | Gene names | Snrpd1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SYS_MOUSE
|
||||||
| NC score | 0.017637 (rank : 39) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26638 | Gene names | Sars, Sars1, Sers | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.016902 (rank : 40) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.016843 (rank : 41) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ANKZ1_HUMAN
|
||||||
| NC score | 0.016061 (rank : 42) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8Y5, Q9NVZ4 | Gene names | ANKZF1, ZNF744 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1 (Zinc finger protein 744). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.015580 (rank : 43) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.013092 (rank : 44) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.010934 (rank : 45) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.010589 (rank : 46) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.008404 (rank : 47) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ATS18_HUMAN
|
||||||
| NC score | 0.007497 (rank : 48) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE60, Q6P4R5, Q6ZWJ9 | Gene names | ADAMTS18, ADAMTS21 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-18 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 18) (ADAM-TS 18) (ADAM-TS18). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.002628 (rank : 49) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.001356 (rank : 50) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.000824 (rank : 51) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||