Please be patient as the page loads
|
PDE3A_MOUSE
|
||||||
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993518 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.983104 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 3.98324e-48 (rank : 4) | NC score | 0.902490 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 1.15895e-47 (rank : 5) | NC score | 0.911762 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 1.97686e-47 (rank : 6) | NC score | 0.911189 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 1.67352e-46 (rank : 7) | NC score | 0.902067 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 8) | NC score | 0.891369 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 9) | NC score | 0.907661 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 10) | NC score | 0.890362 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 7.27602e-42 (rank : 11) | NC score | 0.906620 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 7.27602e-42 (rank : 12) | NC score | 0.890268 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 13) | NC score | 0.889414 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 14) | NC score | 0.892203 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 15) | NC score | 0.892277 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 16) | NC score | 0.889144 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 17) | NC score | 0.886102 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 18) | NC score | 0.883137 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 19) | NC score | 0.883103 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 20) | NC score | 0.885875 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 21) | NC score | 0.885167 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 22) | NC score | 0.873256 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 23) | NC score | 0.876977 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 24) | NC score | 0.880270 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 25) | NC score | 0.878054 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 26) | NC score | 0.794497 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 27) | NC score | 0.793466 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 28) | NC score | 0.768194 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 29) | NC score | 0.767562 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 30) | NC score | 0.732570 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 31) | NC score | 0.722965 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6A_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 32) | NC score | 0.726482 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 33) | NC score | 0.732929 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 34) | NC score | 0.722654 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 35) | NC score | 0.718132 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 36) | NC score | 0.735061 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 37) | NC score | 0.736289 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 38) | NC score | 0.752797 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.035396 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 40) | NC score | 0.017297 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.010038 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.031373 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.031921 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.020092 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.027299 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.031085 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.016141 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.014349 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.004966 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.016990 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | -0.000601 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PCDA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.002382 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.018951 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.006710 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.014329 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.012178 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.022548 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.024676 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.011857 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.008929 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.018601 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.021980 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
GPR97_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.012366 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y34, Q6ZMF4, Q86SL9, Q8IZF1 | Gene names | GPR97, PGR26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor (G-protein coupled receptor PGR26). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.005998 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.007387 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
KPCT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | -0.002500 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.014072 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.005116 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.003721 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MAPK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | -0.000514 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | -0.000459 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.016061 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.016064 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.005612 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.028449 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.012115 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ADA2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | -0.002638 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||
|
CAD26_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.002679 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
GPR97_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.012128 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.006953 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.014713 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.014731 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.006215 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
PRKN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.009328 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
TAF12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.015467 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16514, Q15775 | Gene names | TAF12, TAF15, TAF2J, TAFII20 | |||
|
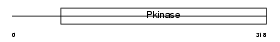
Domain Architecture |
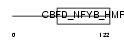
|
|||||
| Description | Transcription initiation factor TFIID subunit 12 (Transcription initiation factor TFIID 20/15 kDa subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.014887 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CTCF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | -0.004578 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008939 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
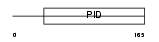
|
|||||
| Description | Disabled homolog 1. | |||||
|
DREB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.007775 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
REPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.007575 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.005804 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.015843 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.003675 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.008711 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FXL19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.009018 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.011718 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.008328 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.005521 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.014024 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.009489 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009058 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.010959 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.001624 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAD26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.003635 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
COTE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010238 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P81408 | Gene names | C1orf2, COTE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein COTE1. | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.003911 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
FOXQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.001453 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70220 | Gene names | Foxq1, Hfh1, Hfh1l | |||
|
Domain Architecture |
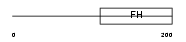
|
|||||
| Description | Forkhead box protein Q1 (Hepatocyte nuclear factor 3 forkhead homolog 1) (HNF-3/forkhead-like protein 1) (HFH-1) (HFH-1l). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.006715 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.003643 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HSN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.007848 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | -0.001700 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.008889 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SNX18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.003463 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.003979 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.004232 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.993518 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.983104 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.911762 (rank : 4) | θ value | 1.15895e-47 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.911189 (rank : 5) | θ value | 1.97686e-47 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.907661 (rank : 6) | θ value | 3.26607e-42 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.906620 (rank : 7) | θ value | 7.27602e-42 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.902490 (rank : 8) | θ value | 3.98324e-48 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.902067 (rank : 9) | θ value | 1.67352e-46 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.892277 (rank : 10) | θ value | 1.6774e-38 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.892203 (rank : 11) | θ value | 4.71619e-41 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.891369 (rank : 12) | θ value | 2.50074e-42 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.890362 (rank : 13) | θ value | 5.57106e-42 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.890268 (rank : 14) | θ value | 7.27602e-42 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.889414 (rank : 15) | θ value | 2.76489e-41 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.889144 (rank : 16) | θ value | 8.32485e-38 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.886102 (rank : 17) | θ value | 2.05049e-36 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.885875 (rank : 18) | θ value | 3.74554e-30 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.885167 (rank : 19) | θ value | 4.89182e-30 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.883137 (rank : 20) | θ value | 1.12514e-34 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.883103 (rank : 21) | θ value | 9.52487e-34 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.880270 (rank : 22) | θ value | 1.73987e-27 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.878054 (rank : 23) | θ value | 3.28125e-26 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.876977 (rank : 24) | θ value | 7.80994e-28 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.873256 (rank : 25) | θ value | 7.80994e-28 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.794497 (rank : 26) | θ value | 2.06002e-20 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.793466 (rank : 27) | θ value | 4.58923e-20 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.768194 (rank : 28) | θ value | 5.07402e-19 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.767562 (rank : 29) | θ value | 1.13037e-18 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.752797 (rank : 30) | θ value | 1.133e-10 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.736289 (rank : 31) | θ value | 4.59992e-12 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.735061 (rank : 32) | θ value | 9.26847e-13 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.732929 (rank : 33) | θ value | 5.8054e-15 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.732570 (rank : 34) | θ value | 2.60593e-15 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.726482 (rank : 35) | θ value | 5.8054e-15 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.722965 (rank : 36) | θ value | 3.40345e-15 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.722654 (rank : 37) | θ value | 7.58209e-15 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.718132 (rank : 38) | θ value | 1.42992e-13 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.035396 (rank : 39) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.031921 (rank : 40) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.031373 (rank : 41) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.031085 (rank : 42) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.028449 (rank : 43) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.027299 (rank : 44) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.024676 (rank : 45) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.022548 (rank : 46) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.021980 (rank : 47) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.020092 (rank : 48) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.018951 (rank : 49) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.018601 (rank : 50) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.017297 (rank : 51) | θ value | 0.0961366 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.016990 (rank : 52) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.016141 (rank : 53) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.016064 (rank : 54) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.016061 (rank : 55) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.015843 (rank : 56) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TAF12_HUMAN
|
||||||
| NC score | 0.015467 (rank : 57) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16514, Q15775 | Gene names | TAF12, TAF15, TAF2J, TAFII20 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 12 (Transcription initiation factor TFIID 20/15 kDa subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.014887 (rank : 58) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.014731 (rank : 59) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.014713 (rank : 60) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.014349 (rank : 61) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.014329 (rank : 62) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.014072 (rank : 63) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.014024 (rank : 64) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
GPR97_HUMAN
|
||||||
| NC score | 0.012366 (rank : 65) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y34, Q6ZMF4, Q86SL9, Q8IZF1 | Gene names | GPR97, PGR26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor (G-protein coupled receptor PGR26). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.012178 (rank : 66) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GPR97_MOUSE
|
||||||
| NC score | 0.012128 (rank : 67) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.012115 (rank : 68) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.011857 (rank : 69) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.011718 (rank : 70) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.010959 (rank : 71) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
COTE1_HUMAN
|
||||||
| NC score | 0.010238 (rank : 72) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P81408 | Gene names | C1orf2, COTE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein COTE1. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.010038 (rank : 73) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.009489 (rank : 74) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
PRKN2_HUMAN
|
||||||
| NC score | 0.009328 (rank : 75) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.009058 (rank : 76) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
FXL19_HUMAN
|
||||||
| NC score | 0.009018 (rank : 77) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.008939 (rank : 78) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.008929 (rank : 79) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.008889 (rank : 80) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.008711 (rank : 81) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.008328 (rank : 82) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
HSN2_HUMAN
|
||||||
| NC score | 0.007848 (rank : 83) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IFS5 | Gene names | HSN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.007775 (rank : 84) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.007575 (rank : 85) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.007387 (rank : 86) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.006953 (rank : 87) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.006715 (rank : 88) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.006710 (rank : 89) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.006215 (rank : 90) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.005998 (rank : 91) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.005804 (rank : 92) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.005612 (rank : 93) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.005521 (rank : 94) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.005116 (rank : 95) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.004966 (rank : 96) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.004232 (rank : 97) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.003979 (rank : 98) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.003911 (rank : 99) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.003721 (rank : 100) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.003675 (rank : 101) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.003643 (rank : 102) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
CAD26_MOUSE
|
||||||
| NC score | 0.003635 (rank : 103) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
SNX18_HUMAN
|
||||||
| NC score | 0.003463 (rank : 104) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
CAD26_HUMAN
|
||||||
| NC score | 0.002679 (rank : 105) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
PCDA3_HUMAN
|
||||||
| NC score | 0.002382 (rank : 106) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.001624 (rank : 107) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
FOXQ1_MOUSE
|
||||||
| NC score | 0.001453 (rank : 108) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70220 | Gene names | Foxq1, Hfh1, Hfh1l | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein Q1 (Hepatocyte nuclear factor 3 forkhead homolog 1) (HNF-3/forkhead-like protein 1) (HFH-1) (HFH-1l). | |||||
|
MAPK2_MOUSE
|
||||||
| NC score | -0.000459 (rank : 109) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_HUMAN
|
||||||
| NC score | -0.000514 (rank : 110) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | -0.000601 (rank : 111) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | -0.001700 (rank : 112) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
KPCT_MOUSE
|
||||||
| NC score | -0.002500 (rank : 113) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
ADA2A_HUMAN
|
||||||
| NC score | -0.002638 (rank : 114) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||
|
CTCF_MOUSE
|
||||||
| NC score | -0.004578 (rank : 115) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||