Please be patient as the page loads
|
DAB1_MOUSE
|
||||||
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
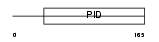
|
|||||
| Description | Disabled homolog 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DAB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.956150 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 3.59435e-57 (rank : 3) | NC score | 0.790774 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
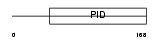
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 5.19021e-56 (rank : 4) | NC score | 0.792199 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
ARH_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 5) | NC score | 0.469636 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 6) | NC score | 0.457616 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 7) | NC score | 0.397747 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 8) | NC score | 0.399509 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 9) | NC score | 0.375798 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 10) | NC score | 0.403349 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.232676 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 12) | NC score | 0.110572 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ANKS1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.115673 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.103519 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.123259 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.147942 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.128555 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.059645 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.137445 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CHM1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.107287 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.072700 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.068304 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.098406 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.103727 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.049527 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
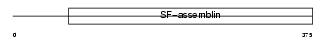
SFPQ_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.068125 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.101301 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.027583 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
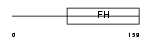
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.016363 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CHM1A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.092749 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.103159 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.101599 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
HEM1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.039543 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
HIPK2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.011159 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
LPIN2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.042079 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |
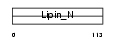
|
|||||
| Description | Lipin-2. | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.020986 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.119753 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.119799 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.070246 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.042172 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.056839 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.083477 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.044296 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.095652 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.040180 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.088260 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.072920 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
DACH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.034039 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.023903 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.100355 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
K0240_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.063954 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.051351 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.076475 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.055803 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.085098 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.087891 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.127037 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
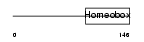
DLX4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.017421 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.020853 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.036711 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.042595 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.016384 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ARRD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.029079 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.013898 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.016181 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.003466 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.039223 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.032762 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.077665 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.074582 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.040239 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.036033 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.034628 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.066911 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.047396 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.011478 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.037462 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.089655 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.036730 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.038276 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AMELY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.062102 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
CASC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.078579 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.080581 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.040522 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.024648 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.033798 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
HGS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.039956 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.099919 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.074049 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.021945 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.044472 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.084221 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.081413 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.042589 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.019295 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.042745 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1C23_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.006117 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PS0 | Gene names | Krt23, Haik1, Krt1-23 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 23 (Cytokeratin-23) (CK-23) (Keratin-23) (K23). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.065086 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PELI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.024478 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C669, Q91YL9, Q9CV22, Q9ERJ8 | Gene names | Peli1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 1 (Pellino-1). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.036226 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.090388 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.049440 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.031511 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
AMBN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.036080 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |
|
|||||
| Description | Ameloblastin precursor. | |||||
|
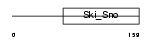
DACH2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.028184 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |
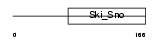
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.015184 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.032466 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.048047 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.063860 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
OBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.091335 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16633, Q14983 | Gene names | POU2AF1, OBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (OCA-B). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.068708 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.113458 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MTF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.001321 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07243 | Gene names | Mtf1, Mtf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.008939 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PELI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.018486 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FA3, Q96SM0, Q9GZY5, Q9HCX0 | Gene names | PELI1, PRISM | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 1 (Pellino-1) (Pellino-related intracellular signaling molecule). | |||||
|
PML_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.015175 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.011702 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.044904 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.017013 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.081200 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BBC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.022411 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ML1 | Gene names | Bbc3, Puma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-binding component 3 (p53 up-regulated modulator of apoptosis). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.017906 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
FZR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.004196 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM11, O75869, Q86U66, Q96NW8, Q9UI96, Q9ULH8, Q9UM10, Q9UNQ1, Q9Y2T8 | Gene names | FZR1, CDH1, FYR, FZR, KIAA1242 | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog) (hCDH1) (CDC20-like protein 1). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.032752 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.016566 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.082831 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.012305 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.017320 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.052767 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.026835 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.025925 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.014607 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.020998 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
TRIP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.022009 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15650, Q96ED7, Q9UKH0 | Gene names | TRIP4 | |||
|
Domain Architecture |
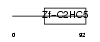
|
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (TRIP-4). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.012689 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ANM5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.012774 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIG8, Q3TNN1, Q9QZS9 | Gene names | Prmt5, Jbp1, Skb1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein arginine N-methyltransferase 5 (EC 2.1.1.125) (EC 2.1.1.-) (Shk1 kinase-binding protein 1 homolog) (SKB1 homolog) (Jak-binding protein 1). | |||||
|
CASL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.019978 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
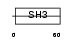
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.013549 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.012778 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
HDAC9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.012454 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.019900 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
NUP62_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.020685 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.055836 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.074304 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SSA27_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.031983 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60232 | Gene names | SSSCA1 | |||
|
Domain Architecture |
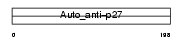
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 (Autoantigen p27). | |||||
|
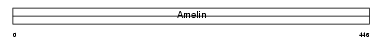
AMELX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.063671 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.076376 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.072281 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.056899 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CAPON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.102826 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
CAPON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.100923 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.068610 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FA43A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.061082 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FA43A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.061038 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUP8, Q8R231 | Gene names | Fam43a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.064240 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.050015 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.058810 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.062296 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.066320 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.074351 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.089951 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.082700 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.056701 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.058789 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.079996 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.056456 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
WASF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051549 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.076917 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.058354 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.052400 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.056603 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.956150 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.792199 (rank : 3) | θ value | 5.19021e-56 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.790774 (rank : 4) | θ value | 3.59435e-57 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
ARH_HUMAN
|
||||||
| NC score | 0.469636 (rank : 5) | θ value | 2.13673e-09 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| NC score | 0.457616 (rank : 6) | θ value | 2.79066e-09 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.403349 (rank : 7) | θ value | 3.41135e-07 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.399509 (rank : 8) | θ value | 1.53129e-07 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.397747 (rank : 9) | θ value | 1.53129e-07 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.375798 (rank : 10) | θ value | 3.41135e-07 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.232676 (rank : 11) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.147942 (rank : 12) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.137445 (rank : 13) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.128555 (rank : 14) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.127037 (rank : 15) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.123259 (rank : 16) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.119799 (rank : 17) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.119753 (rank : 18) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
ANKS1_MOUSE
|
||||||
| NC score | 0.115673 (rank : 19) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.113458 (rank : 20) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.110572 (rank : 21) | θ value | 0.00102713 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
CHM1A_HUMAN
|
||||||
| NC score | 0.107287 (rank : 22) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.103727 (rank : 23) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.103519 (rank : 24) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.103159 (rank : 25) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CAPON_HUMAN
|
||||||
| NC score | 0.102826 (rank : 26) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.101599 (rank : 27) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.101301 (rank : 28) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
CAPON_MOUSE
|
||||||
| NC score | 0.100923 (rank : 29) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.100355 (rank : 30) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.099919 (rank : 31) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.098406 (rank : 32) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.095652 (rank : 33) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
CHM1A_MOUSE
|
||||||
| NC score | 0.092749 (rank : 34) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
OBF1_HUMAN
|
||||||
| NC score | 0.091335 (rank : 35) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16633, Q14983 | Gene names | POU2AF1, OBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (OCA-B). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.090388 (rank : 36) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.089951 (rank : 37) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.089655 (rank : 38) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.088260 (rank : 39) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.087891 (rank : 40) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.085098 (rank : 41) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.084221 (rank : 42) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.083477 (rank : 43) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.082831 (rank : 44) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.082700 (rank : 45) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.081413 (rank : 46) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.081200 (rank : 47) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.080581 (rank : 48) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.079996 (rank : 49) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CASC3_HUMAN
|
||||||
| NC score | 0.078579 (rank : 50) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.077665 (rank : 51) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.076917 (rank : 52) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.076475 (rank : 53) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.076376 (rank : 54) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.074582 (rank : 55) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.074351 (rank : 56) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.074304 (rank : 57) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.074049 (rank : 58) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.072920 (rank : 59) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.072700 (rank : 60) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.072281 (rank : 61) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.070246 (rank : 62) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.068708 (rank : 63) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.068610 (rank : 64) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.068304 (rank : 65) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.068125 (rank : 66) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.066911 (rank : 67) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.066320 (rank : 68) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.065086 (rank : 69) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.064240 (rank : 70) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.063954 (rank : 71) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.063860 (rank : 72) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.063671 (rank : 73) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.062296 (rank : 74) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
AMELY_HUMAN
|
||||||
| NC score | 0.062102 (rank : 75) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
FA43A_HUMAN
|
||||||
| NC score | 0.061082 (rank : 76) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FA43A_MOUSE
|
||||||
| NC score | 0.061038 (rank : 77) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUP8, Q8R231 | Gene names | Fam43a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.059645 (rank : 78) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058810 (rank : 79) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.058789 (rank : 80) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.058354 (rank : 81) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.056899 (rank : 82) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.056839 (rank : 83) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.056701 (rank : 84) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.056603 (rank : 85) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.056456 (rank : 86) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.055836 (rank : 87) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.055803 (rank : 88) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.052767 (rank : 89) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.052400 (rank : 90) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASF1_HUMAN
|
||||||
| NC score | 0.051549 (rank : 91) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.051351 (rank : 92) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.050015 (rank : 93) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.049527 (rank : 94) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.049440 (rank : 95) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.048047 (rank : 96) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.047396 (rank : 97) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.044904 (rank : 98) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.044472 (rank : 99) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.044296 (rank : 100) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.042745 (rank : 101) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.042595 (rank : 102) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.042589 (rank : 103) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.042172 (rank : 104) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
LPIN2_MOUSE
|
||||||
| NC score | 0.042079 (rank : 105) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-2. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.040522 (rank : 106) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.040239 (rank : 107) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.040180 (rank : 108) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.039956 (rank : 109) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HEM1_MOUSE
|
||||||
| NC score | 0.039543 (rank : 110) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.039223 (rank : 111) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.038276 (rank : 112) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.037462 (rank : 113) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.036730 (rank : 114) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.036711 (rank : 115) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.036226 (rank : 116) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
AMBN_HUMAN
|
||||||
| NC score | 0.036080 (rank : 117) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |

|
|||||
| Description | Ameloblastin precursor. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.036033 (rank : 118) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.034628 (rank : 119) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
DACH2_HUMAN
|
||||||
| NC score | 0.034039 (rank : 120) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.033798 (rank : 121) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.032762 (rank : 122) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.032752 (rank : 123) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.032466 (rank : 124) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SSA27_HUMAN
|
||||||
| NC score | 0.031983 (rank : 125) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60232 | Gene names | SSSCA1 | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 (Autoantigen p27). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.031511 (rank : 126) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ARRD1_MOUSE
|
||||||
| NC score | 0.029079 (rank : 127) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
DACH2_MOUSE
|
||||||
| NC score | 0.028184 (rank : 128) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.027583 (rank : 129) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.026835 (rank : 130) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.025925 (rank : 131) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.024648 (rank : 132) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
PELI1_MOUSE
|
||||||
| NC score | 0.024478 (rank : 133) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C669, Q91YL9, Q9CV22, Q9ERJ8 | Gene names | Peli1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 1 (Pellino-1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.023903 (rank : 134) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
BBC3_MOUSE
|
||||||
| NC score | 0.022411 (rank : 135) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ML1 | Gene names | Bbc3, Puma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-binding component 3 (p53 up-regulated modulator of apoptosis). | |||||
|
TRIP4_HUMAN
|
||||||
| NC score | 0.022009 (rank : 136) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15650, Q96ED7, Q9UKH0 | Gene names | TRIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (TRIP-4). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.021945 (rank : 137) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.020998 (rank : 138) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.020986 (rank : 139) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.020853 (rank : 140) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.020685 (rank : 141) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.019978 (rank : 142) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.019900 (rank : 143) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.019295 (rank : 144) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
PELI1_HUMAN
|
||||||
| NC score | 0.018486 (rank : 145) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FA3, Q96SM0, Q9GZY5, Q9HCX0 | Gene names | PELI1, PRISM | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 1 (Pellino-1) (Pellino-related intracellular signaling molecule). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.017906 (rank : 146) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
DLX4_MOUSE
|
||||||
| NC score | 0.017421 (rank : 147) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70436, Q3UM51, Q8R4I3 | Gene names | Dlx4, Dlx7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-4 (DLX-7). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.017320 (rank : 148) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.017013 (rank : 149) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.016566 (rank : 150) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.016384 (rank : 151) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.016363 (rank : 152) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.016181 (rank : 153) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.015184 (rank : 154) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
PML_HUMAN
|
||||||
| NC score | 0.015175 (rank : 155) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.014607 (rank : 156) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.013898 (rank : 157) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.013549 (rank : 158) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.012778 (rank : 159) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
ANM5_MOUSE
|
||||||
| NC score | 0.012774 (rank : 160) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIG8, Q3TNN1, Q9QZS9 | Gene names | Prmt5, Jbp1, Skb1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein arginine N-methyltransferase 5 (EC 2.1.1.125) (EC 2.1.1.-) (Shk1 kinase-binding protein 1 homolog) (SKB1 homolog) (Jak-binding protein 1). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.012689 (rank : 161) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
HDAC9_HUMAN
|
||||||
| NC score | 0.012454 (rank : 162) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.012305 (rank : 163) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.011702 (rank : 164) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.011478 (rank : 165) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
HIPK2_MOUSE
|
||||||
| NC score | 0.011159 (rank : 166) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.008939 (rank : 167) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
K1C23_MOUSE
|
||||||
| NC score | 0.006117 (rank : 168) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PS0 | Gene names | Krt23, Haik1, Krt1-23 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 23 (Cytokeratin-23) (CK-23) (Keratin-23) (K23). | |||||
|
FZR_HUMAN
|
||||||
| NC score | 0.004196 (rank : 169) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UM11, O75869, Q86U66, Q96NW8, Q9UI96, Q9ULH8, Q9UM10, Q9UNQ1, Q9Y2T8 | Gene names | FZR1, CDH1, FYR, FZR, KIAA1242 | |||
|
Domain Architecture |

|
|||||
| Description | Fizzy-related protein homolog (Fzr) (Cdh1/Hct1 homolog) (hCDH1) (CDC20-like protein 1). | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | 0.003466 (rank : 170) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
MTF1_MOUSE
|
||||||
| NC score | 0.001321 (rank : 171) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07243 | Gene names | Mtf1, Mtf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||