Please be patient as the page loads
|
CHM1A_MOUSE
|
||||||
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHM1A_MOUSE
|
||||||
| θ value | 6.71607e-88 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
CHM1A_HUMAN
|
||||||
| θ value | 1.40038e-85 (rank : 2) | NC score | 0.997391 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
CH1B1_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 3) | NC score | 0.950351 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
CHM1B_HUMAN
|
||||||
| θ value | 7.03111e-45 (rank : 4) | NC score | 0.948469 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7LBR1, Q96E89, Q9HD41 | Gene names | CHMP1B, C18orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b (Chromatin-modifying protein 1b) (CHMP1b) (CHMP1.5) (Vacuolar protein sorting 46-2) (Vps46-2) (hVps46-2). | |||||
|
CH1B2_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 5) | NC score | 0.948480 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQD4 | Gene names | Chmp1b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-2 (Chromatin-modifying protein 1b-2) (CHMP1b-2). | |||||
|
CHM2A_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 6) | NC score | 0.659598 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43633, Q3ZTT0 | Gene names | CHMP2A, BC2, CHMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2a (Chromatin-modifying protein 2a) (CHMP2a) (Vacuolar protein sorting 2-1) (Vps2-1) (hVps2-1) (Putative breast adenocarcinoma marker BC-2). | |||||
|
CHM2A_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 7) | NC score | 0.659060 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB34 | Gene names | Chmp2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2a (Chromatin-modifying protein 2a) (CHMP2a) (Vacuolar protein sorting 2) (mVps2). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.089419 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.092749 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
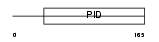
|
|||||
| Description | Disabled homolog 1. | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.043729 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.024074 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.034367 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.040868 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.025781 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.009818 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.030783 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CG301_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.039503 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |
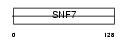
|
|||||
| Description | Protein CGI-301. | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.025867 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.025704 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.023250 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.015991 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.017860 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.012658 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.017439 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CHM4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.037619 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
LRCH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.008001 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.013760 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.021052 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.021966 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
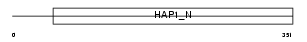
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
CHM4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.035311 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |
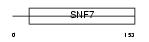
|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.021000 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.008228 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
WDR48_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.003952 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH57, Q80TD4, Q80XI0, Q8BRM0, Q8CBK0, Q922Z9, Q9CRR1, Q9CSL0 | Gene names | Wdr48, Kiaa1449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 48. | |||||
|
CHM2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.221321 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQN3, Q53HC7, Q9Y4U6 | Gene names | CHMP2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2b (Chromatin-modifying protein 2b) (CHMP2b) (CHMP2.5) (Vacuolar protein sorting 2-2) (Vps2-2) (hVps2- 2). | |||||
|
CHM2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.229516 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BJF9, Q80UZ4, Q9CT65 | Gene names | Chmp2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2b (Chromatin-modifying protein 2b) (CHMP2b). | |||||
|
CHMP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.196928 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3E7, Q53S71, Q53SU5, Q9NZ51 | Gene names | VPS24, CHMP3, NEDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 3 (Chromatin-modifying protein 3) (Vacuolar protein sorting-associated protein 24) (hVps24) (Neuroendocrine differentiation factor). | |||||
|
CHMP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.208432 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ10, Q9D2Z2, Q9D7A5 | Gene names | Vps24, Chmp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 3 (Chromatin-modifying protein 3) (Vacuolar protein sorting-associated protein 24). | |||||
|
CHM1A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.71607e-88 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q921W0, Q3TW57 | Gene names | Pcoln3, Chmp1, Chmp1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a). | |||||
|
CHM1A_HUMAN
|
||||||
| NC score | 0.997391 (rank : 2) | θ value | 1.40038e-85 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HD42, Q14468, Q15779, Q96G31 | Gene names | PCOLN3, CHMP1, CHMP1A, KIAA0047, PRSM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1a (Chromatin-modifying protein 1a) (CHMP1a) (Vacuolar protein sorting 46-1) (Vps46-1) (hVps46-1). | |||||
|
CH1B1_MOUSE
|
||||||
| NC score | 0.950351 (rank : 3) | θ value | 1.08474e-45 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
CH1B2_MOUSE
|
||||||
| NC score | 0.948480 (rank : 4) | θ value | 1.19932e-44 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQD4 | Gene names | Chmp1b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-2 (Chromatin-modifying protein 1b-2) (CHMP1b-2). | |||||
|
CHM1B_HUMAN
|
||||||
| NC score | 0.948469 (rank : 5) | θ value | 7.03111e-45 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7LBR1, Q96E89, Q9HD41 | Gene names | CHMP1B, C18orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b (Chromatin-modifying protein 1b) (CHMP1b) (CHMP1.5) (Vacuolar protein sorting 46-2) (Vps46-2) (hVps46-2). | |||||
|
CHM2A_HUMAN
|
||||||
| NC score | 0.659598 (rank : 6) | θ value | 1.09485e-13 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43633, Q3ZTT0 | Gene names | CHMP2A, BC2, CHMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2a (Chromatin-modifying protein 2a) (CHMP2a) (Vacuolar protein sorting 2-1) (Vps2-1) (hVps2-1) (Putative breast adenocarcinoma marker BC-2). | |||||
|
CHM2A_MOUSE
|
||||||
| NC score | 0.659060 (rank : 7) | θ value | 1.09485e-13 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB34 | Gene names | Chmp2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2a (Chromatin-modifying protein 2a) (CHMP2a) (Vacuolar protein sorting 2) (mVps2). | |||||
|
CHM2B_MOUSE
|
||||||
| NC score | 0.229516 (rank : 8) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BJF9, Q80UZ4, Q9CT65 | Gene names | Chmp2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2b (Chromatin-modifying protein 2b) (CHMP2b). | |||||
|
CHM2B_HUMAN
|
||||||
| NC score | 0.221321 (rank : 9) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQN3, Q53HC7, Q9Y4U6 | Gene names | CHMP2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 2b (Chromatin-modifying protein 2b) (CHMP2b) (CHMP2.5) (Vacuolar protein sorting 2-2) (Vps2-2) (hVps2- 2). | |||||
|
CHMP3_MOUSE
|
||||||
| NC score | 0.208432 (rank : 10) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ10, Q9D2Z2, Q9D7A5 | Gene names | Vps24, Chmp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 3 (Chromatin-modifying protein 3) (Vacuolar protein sorting-associated protein 24). | |||||
|
CHMP3_HUMAN
|
||||||
| NC score | 0.196928 (rank : 11) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3E7, Q53S71, Q53SU5, Q9NZ51 | Gene names | VPS24, CHMP3, NEDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 3 (Chromatin-modifying protein 3) (Vacuolar protein sorting-associated protein 24) (hVps24) (Neuroendocrine differentiation factor). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.092749 (rank : 12) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.089419 (rank : 13) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.043729 (rank : 14) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.040868 (rank : 15) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
CG301_HUMAN
|
||||||
| NC score | 0.039503 (rank : 16) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CGI-301. | |||||
|
CHM4B_MOUSE
|
||||||
| NC score | 0.037619 (rank : 17) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CHM4B_HUMAN
|
||||||
| NC score | 0.035311 (rank : 18) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.034367 (rank : 19) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.030783 (rank : 20) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.025867 (rank : 21) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.025781 (rank : 22) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.025704 (rank : 23) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.024074 (rank : 24) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.023250 (rank : 25) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.021966 (rank : 26) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.021052 (rank : 27) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.021000 (rank : 28) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.017860 (rank : 29) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.017439 (rank : 30) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.015991 (rank : 31) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.013760 (rank : 32) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.012658 (rank : 33) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.009818 (rank : 34) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.008228 (rank : 35) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
LRCH2_MOUSE
|
||||||
| NC score | 0.008001 (rank : 36) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
WDR48_MOUSE
|
||||||
| NC score | 0.003952 (rank : 37) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH57, Q80TD4, Q80XI0, Q8BRM0, Q8CBK0, Q922Z9, Q9CRR1, Q9CSL0 | Gene names | Wdr48, Kiaa1449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 48. | |||||